Barcode count pre-processing
Max Trauernicht
Netherlands Cancer Institute - van Steensel labm.trauernicht@nki.nl
14/06/2023
Aim
pMT06 (the pDNA library) was transfected into MCF7 cells that are either TP53 proficient or TP53-KO - 24 hours later mRNA was isolated and barcodes were quantified by sequencing together with pMT06 pDNA counts. This was done in three independent replicates. In this script the barcode counts will be analyzed and some quality checks will be done.
Setup
Libraries
Functions
Loading data
Creating count data frames
Read distribution
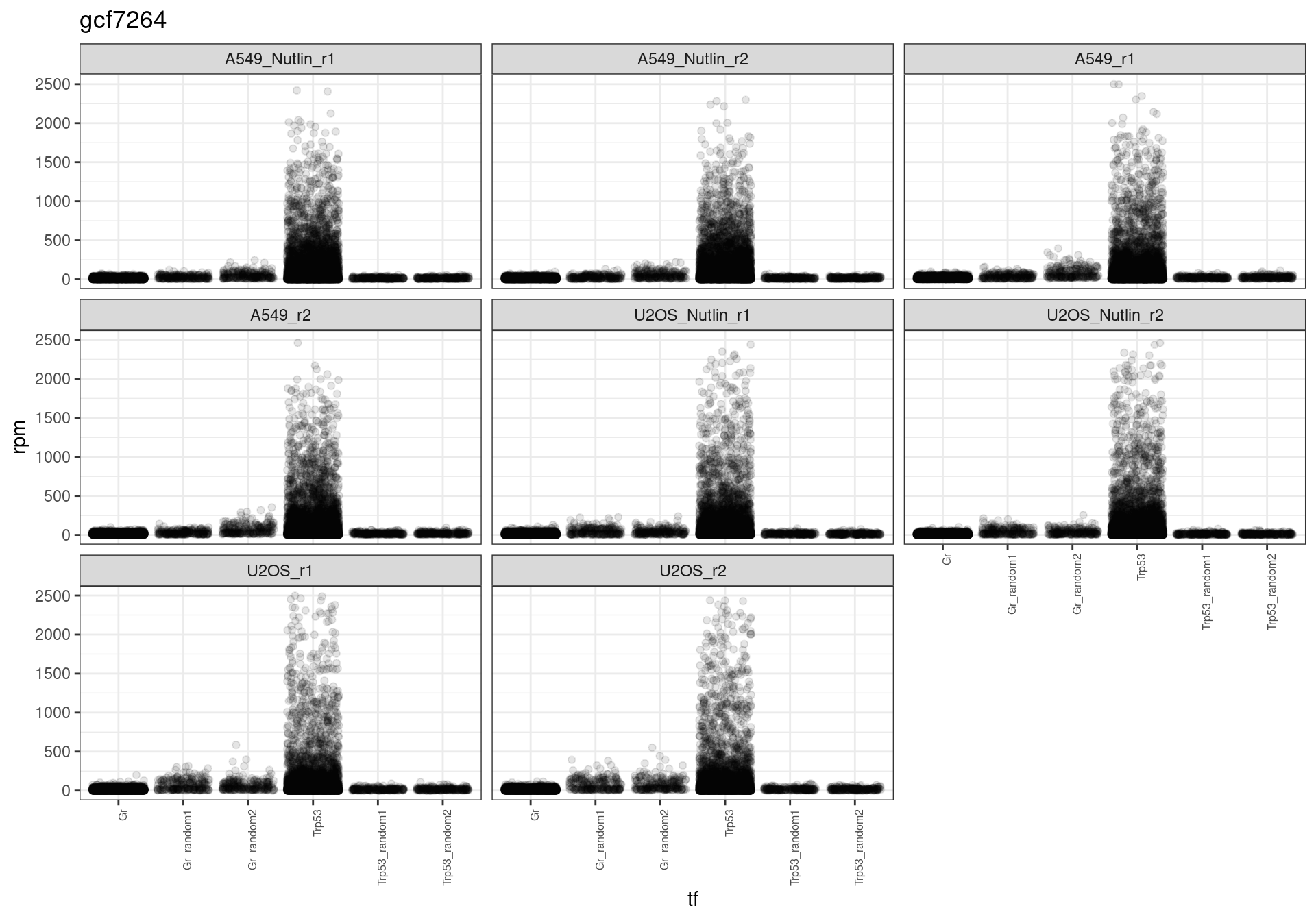
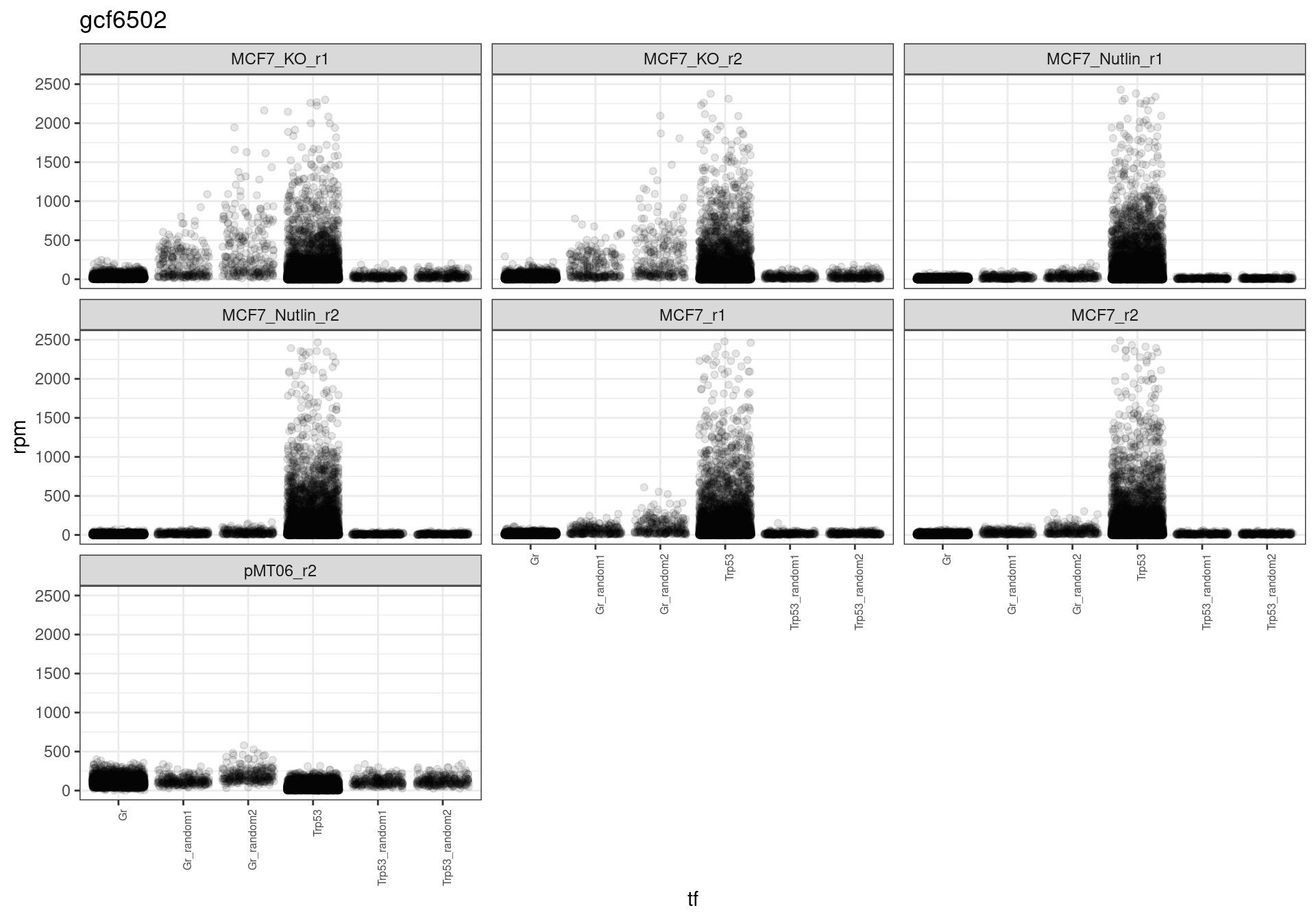
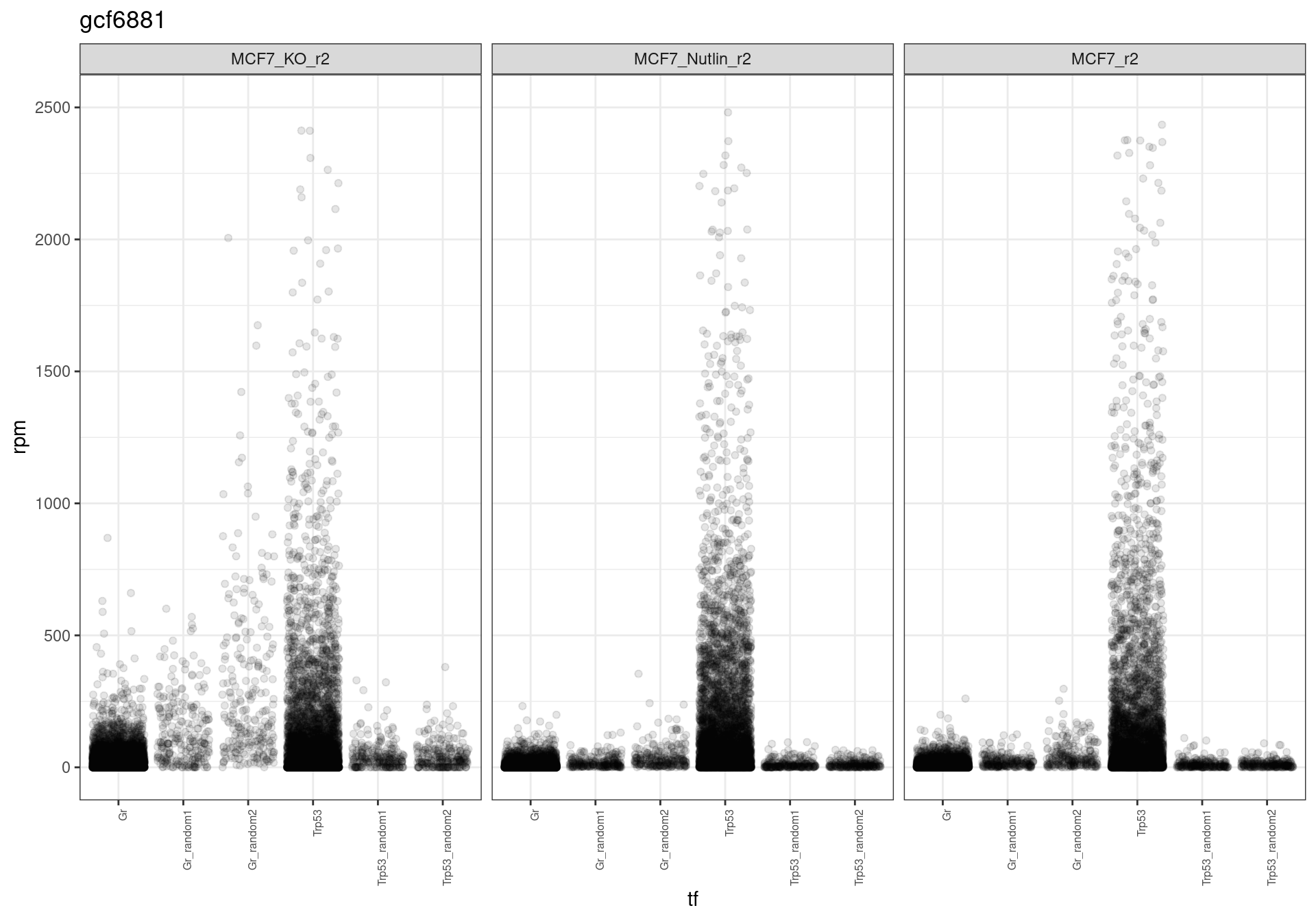
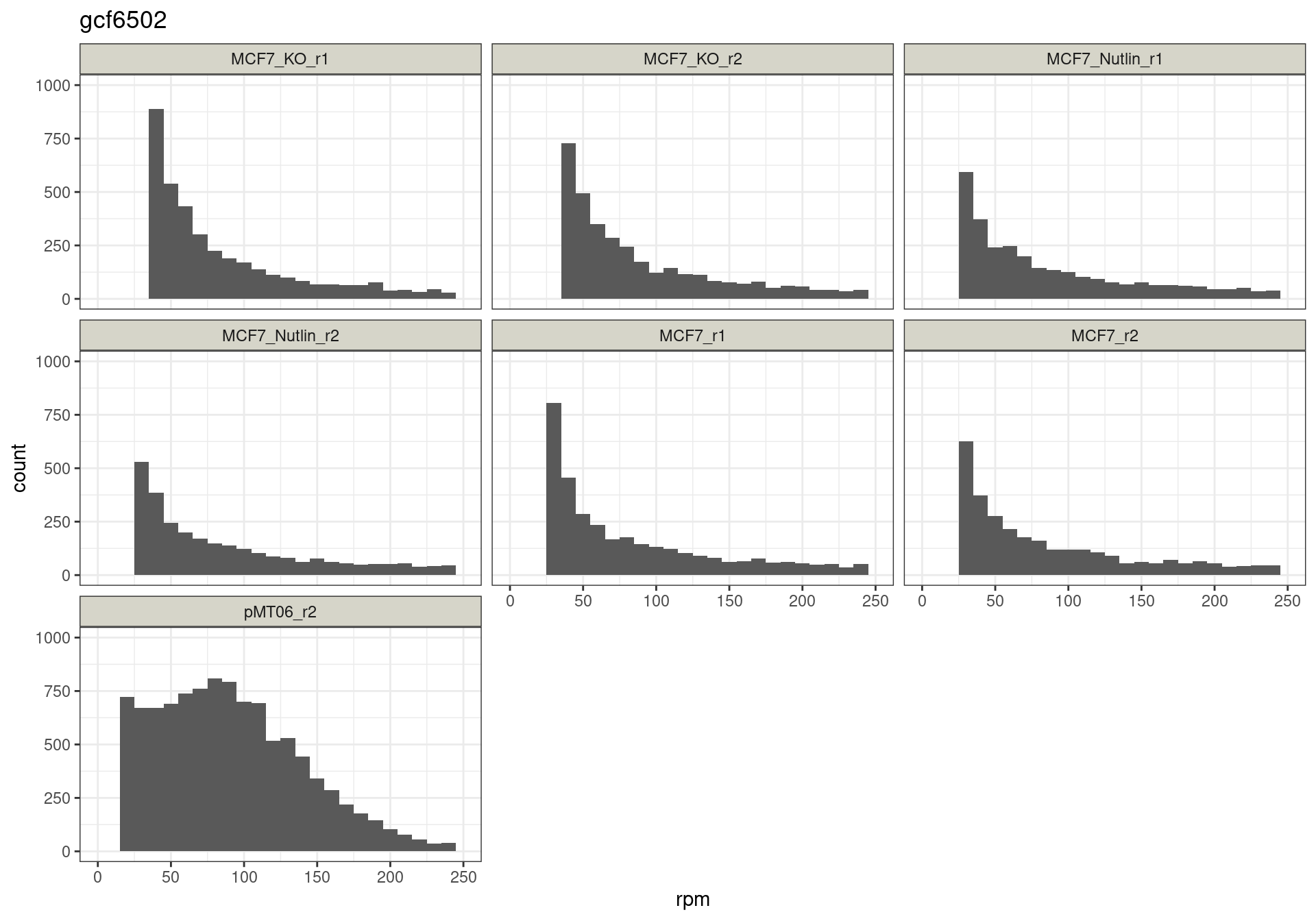
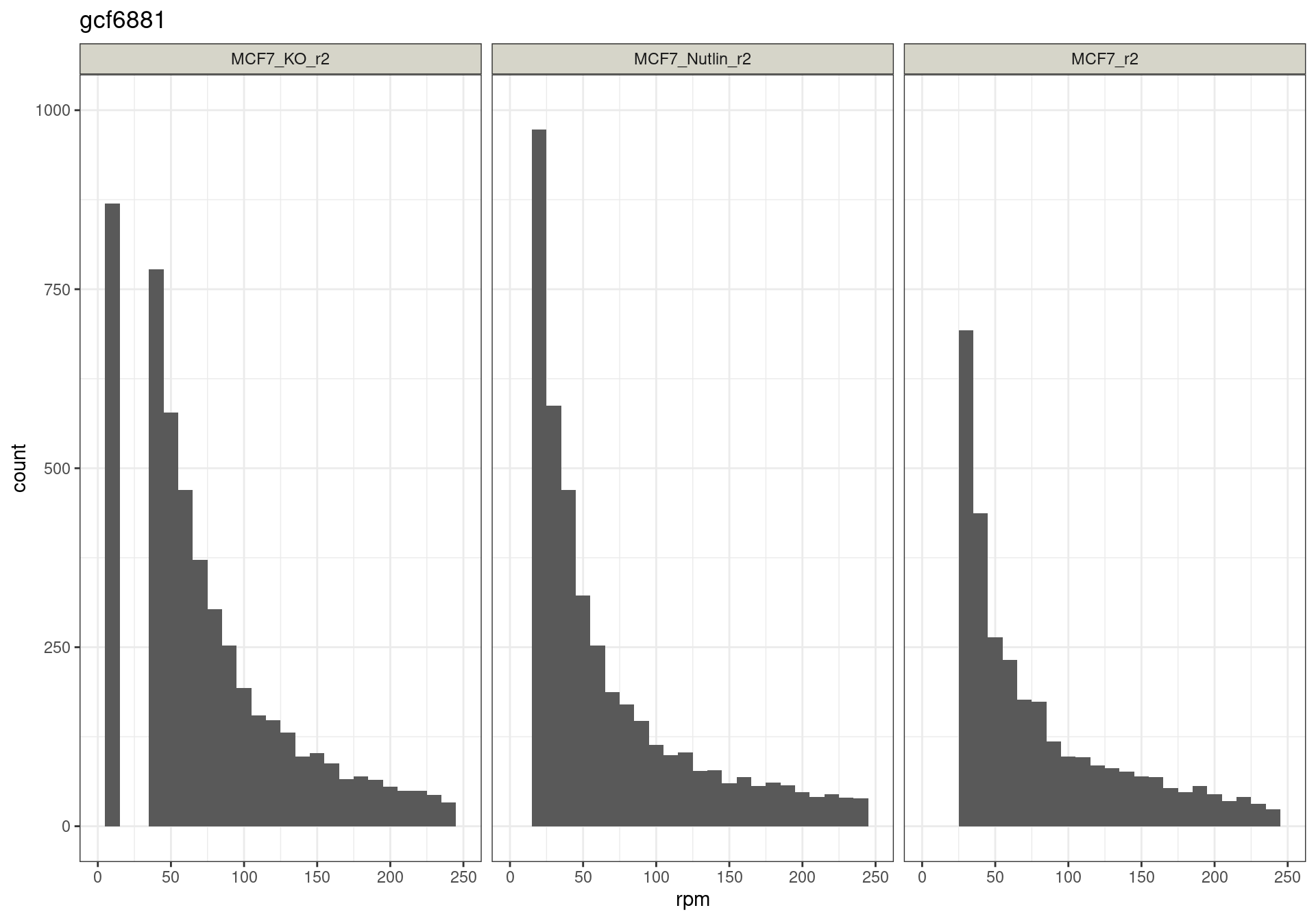
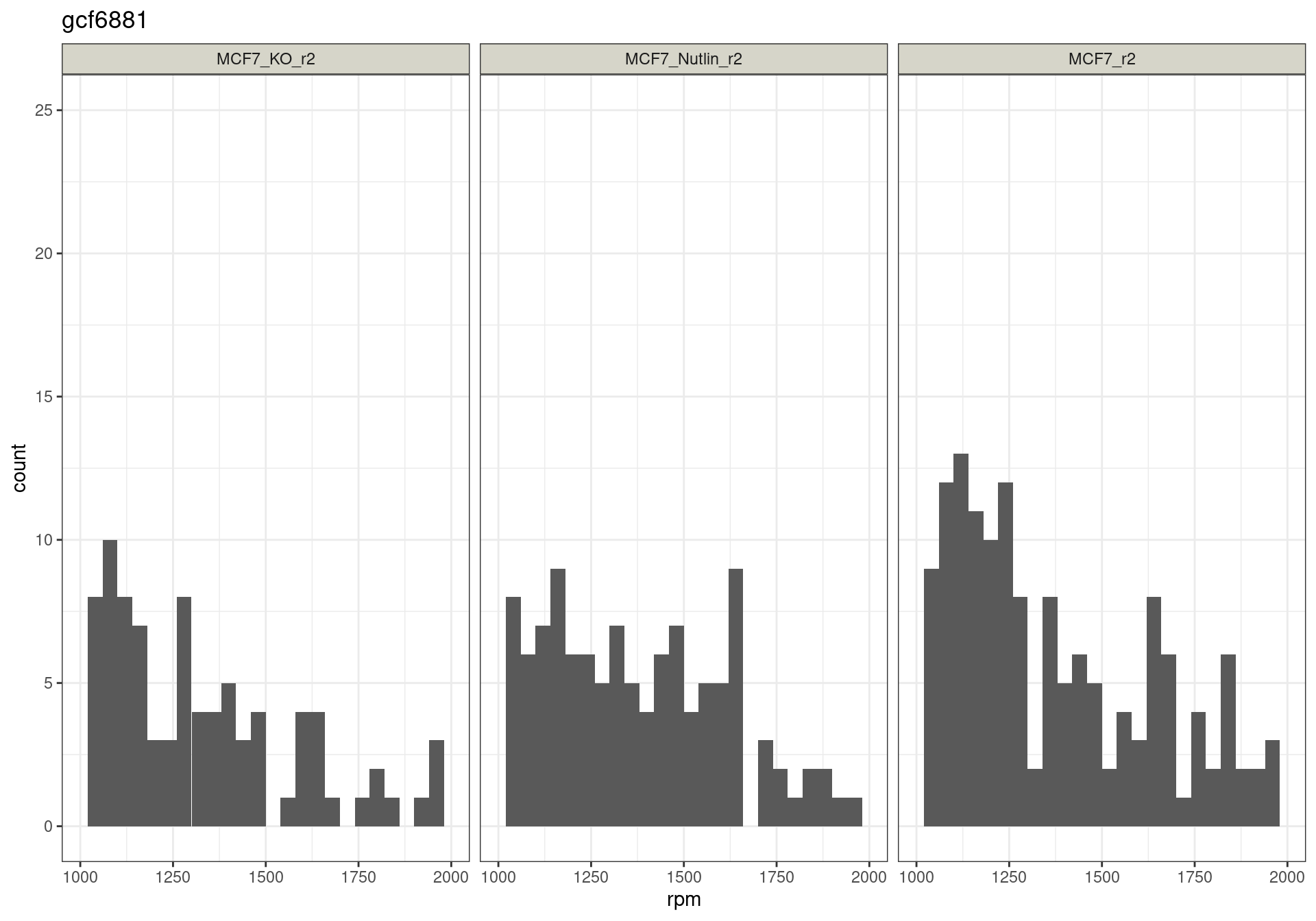
The read distribution plots show that the pDNA samples are uniformly distributed. They also show that the MCF7-TP53-WT cells have highly active TP53 reporters, while random reporters got a lot of reads in the MCF7-TP53-KO cells.
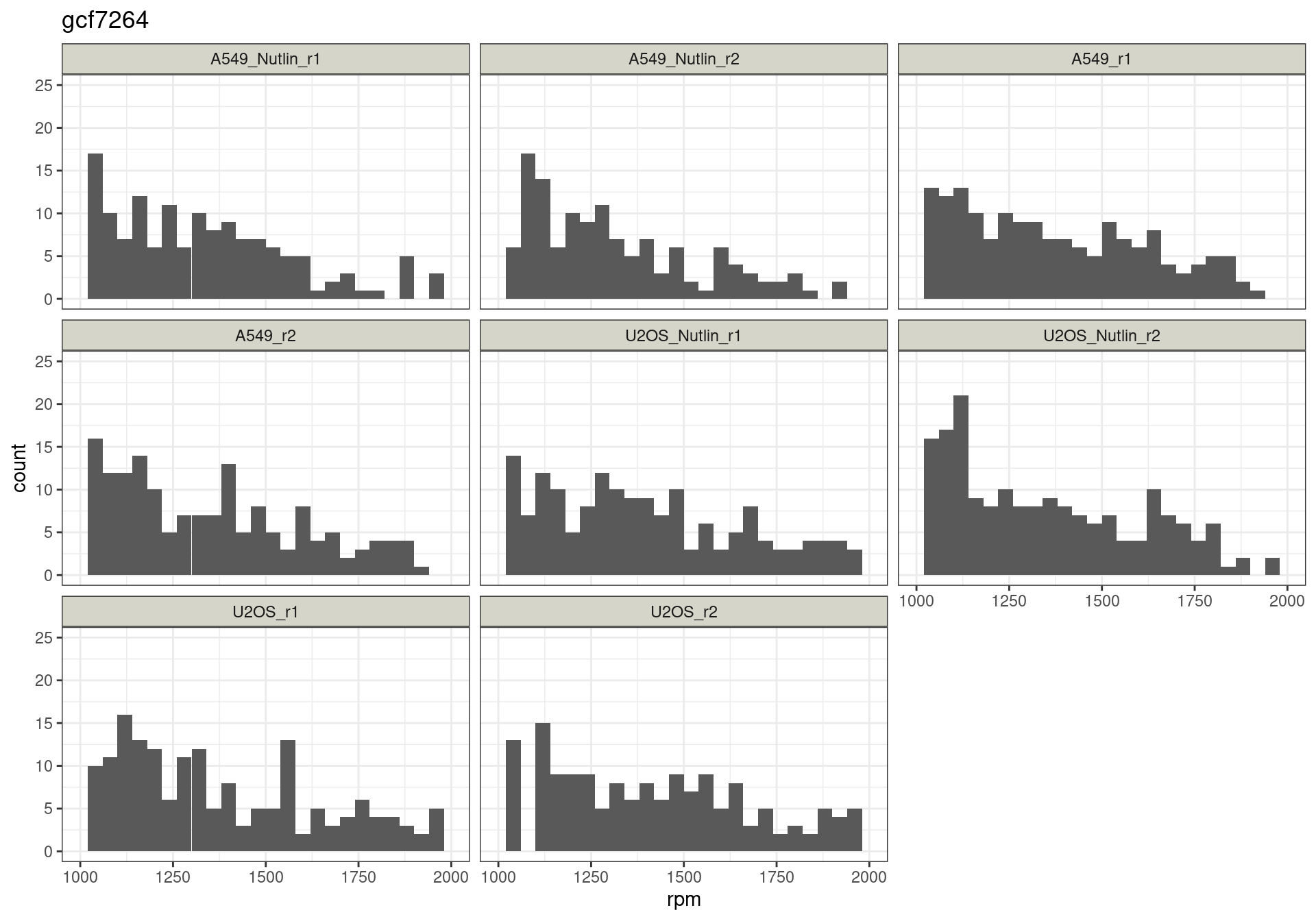
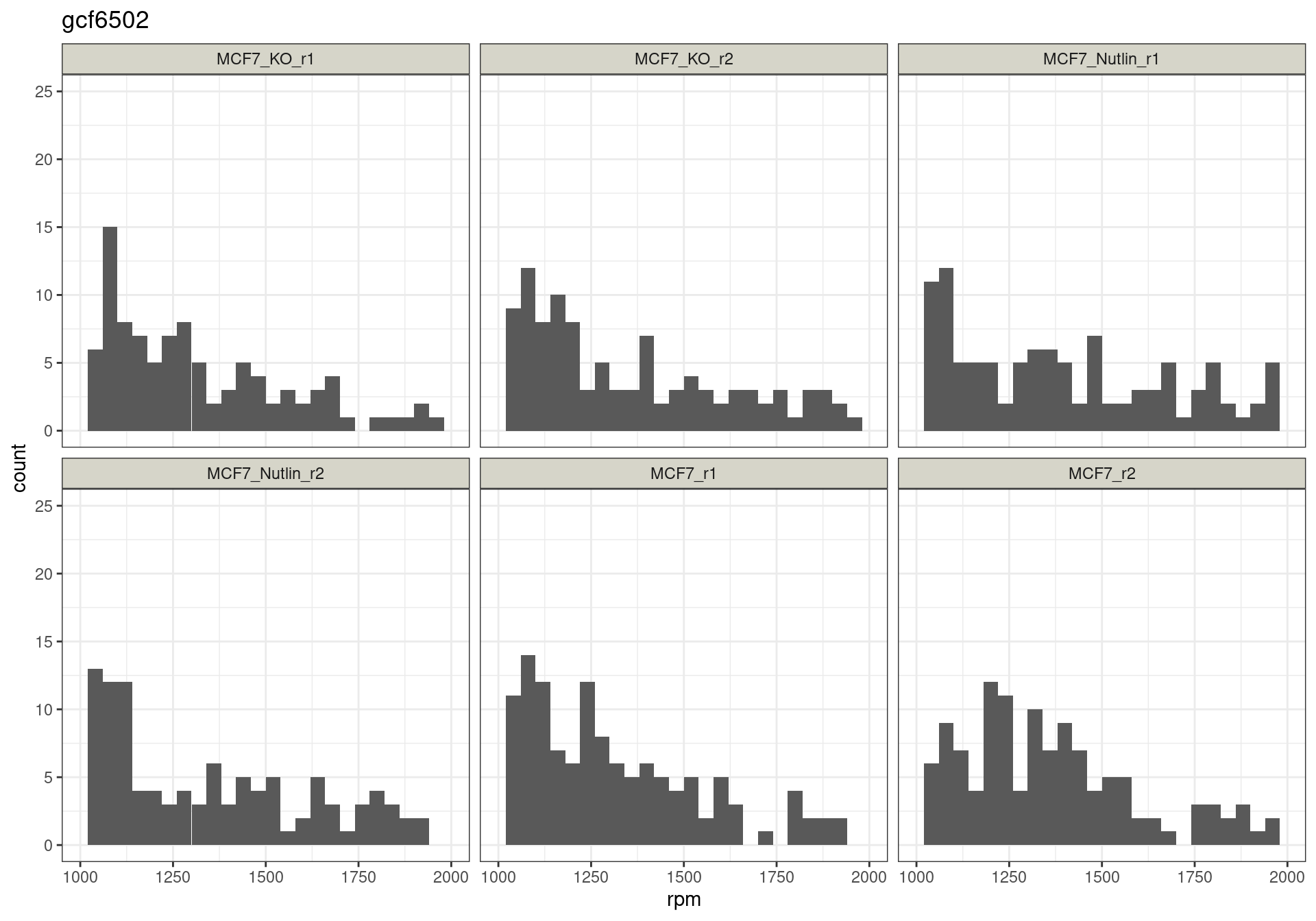
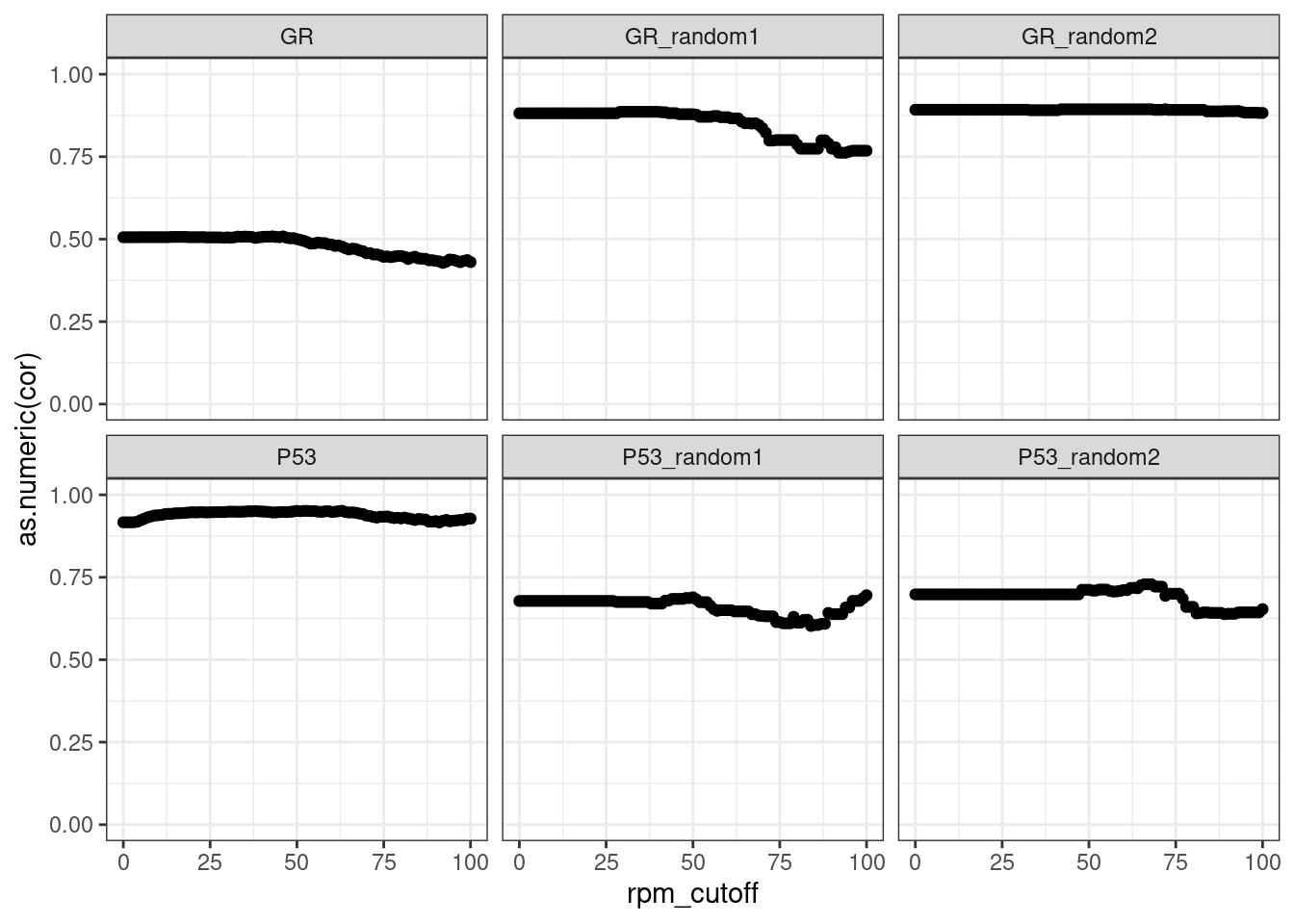
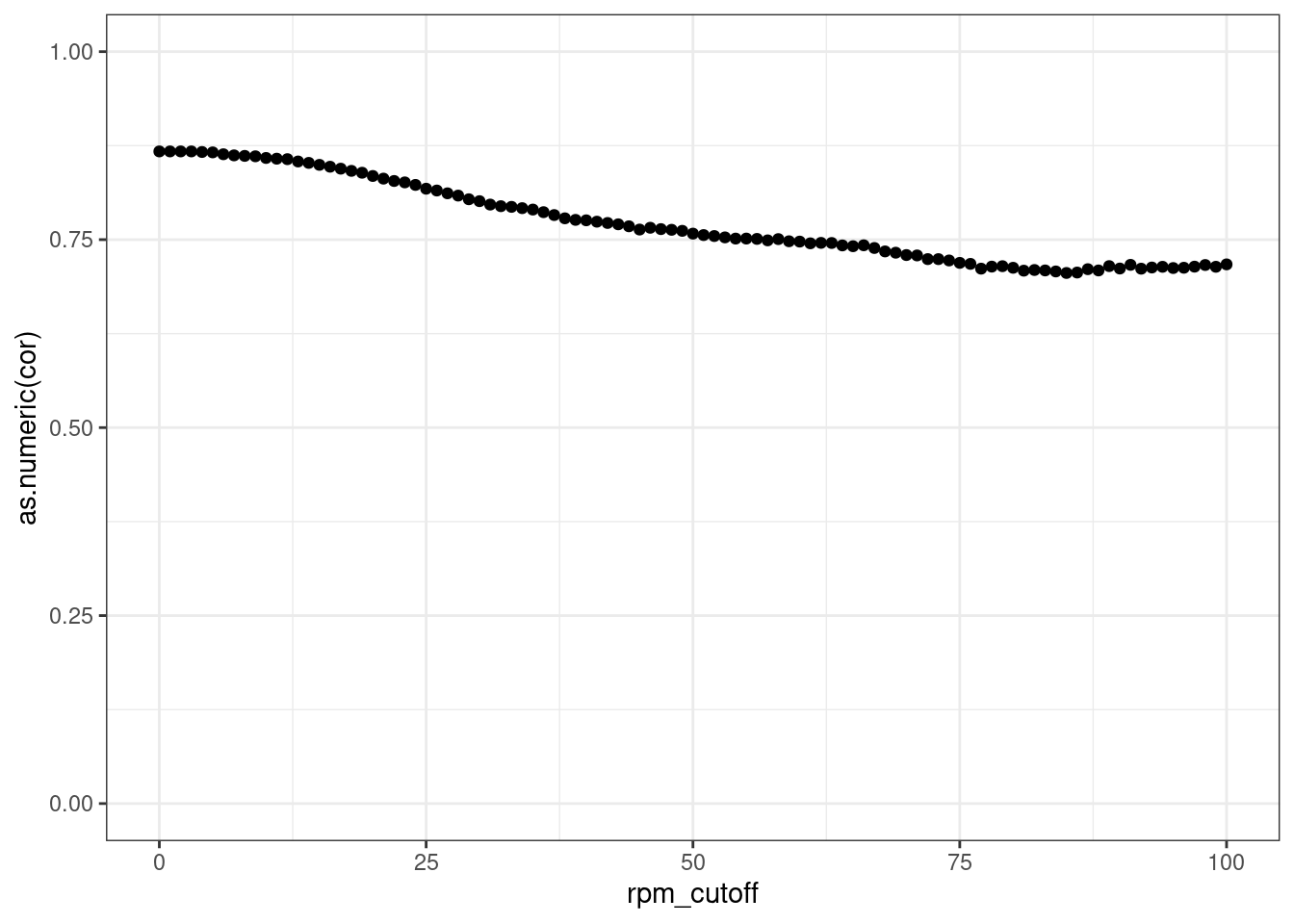
Read distribution per cutoff
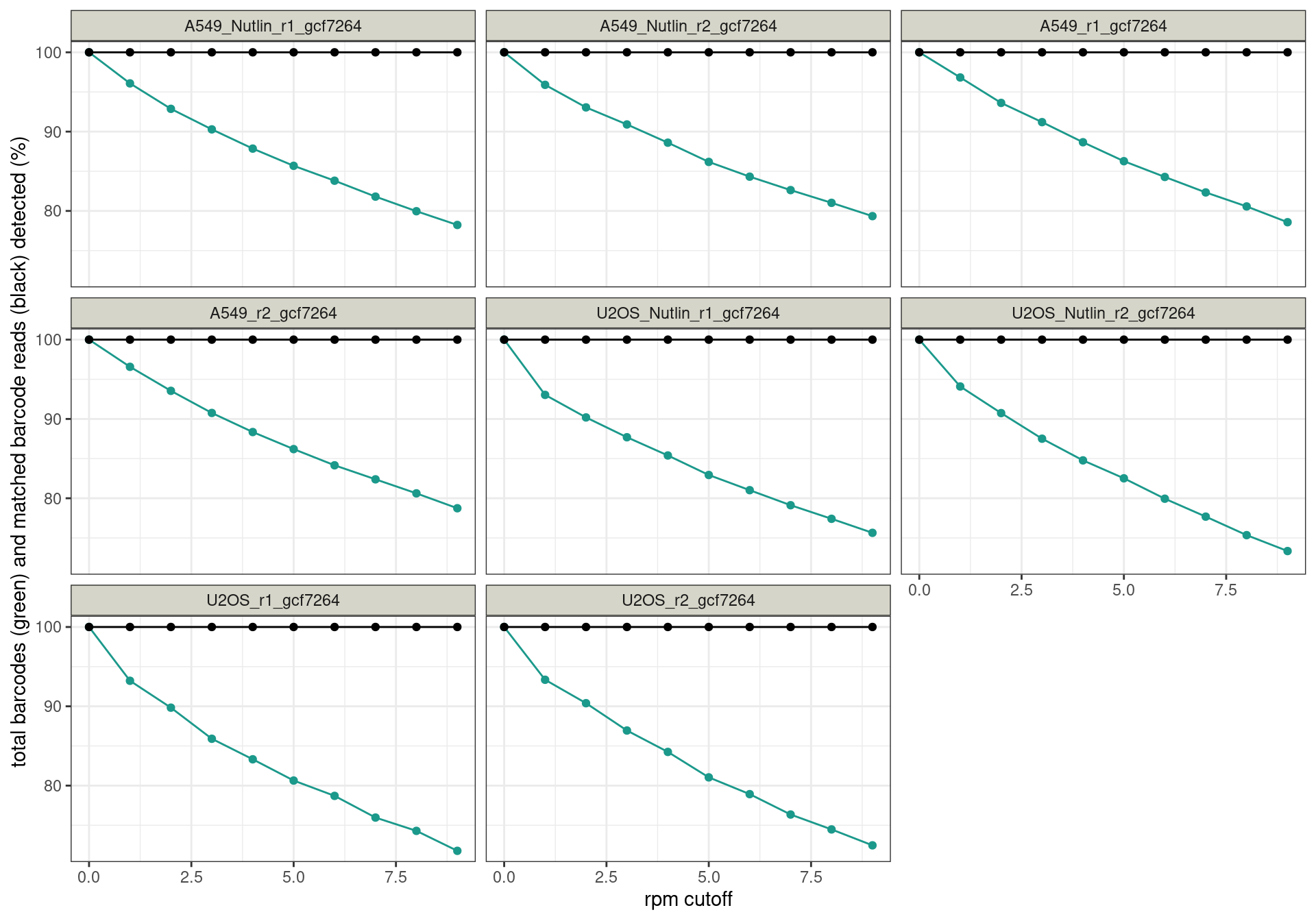
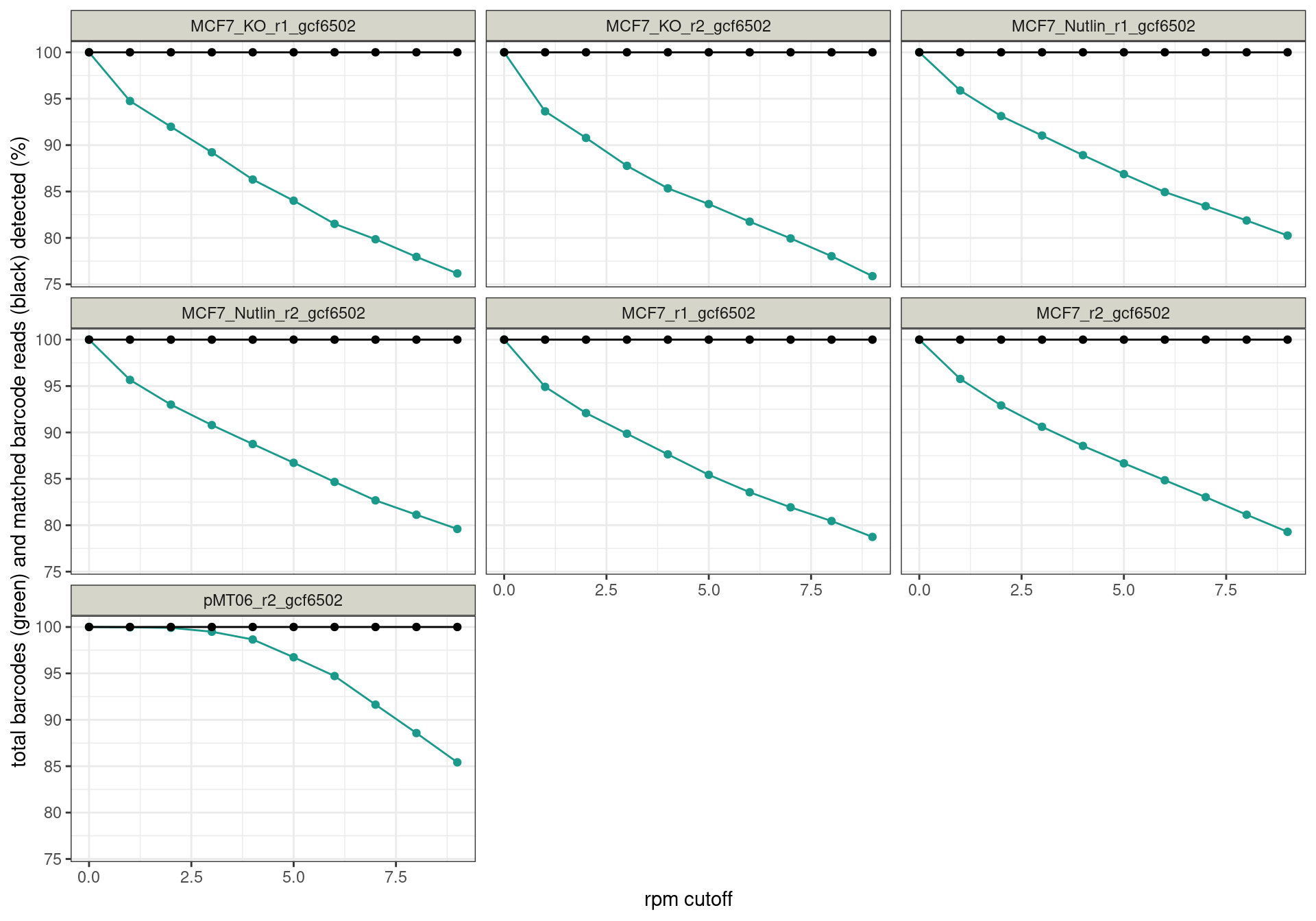
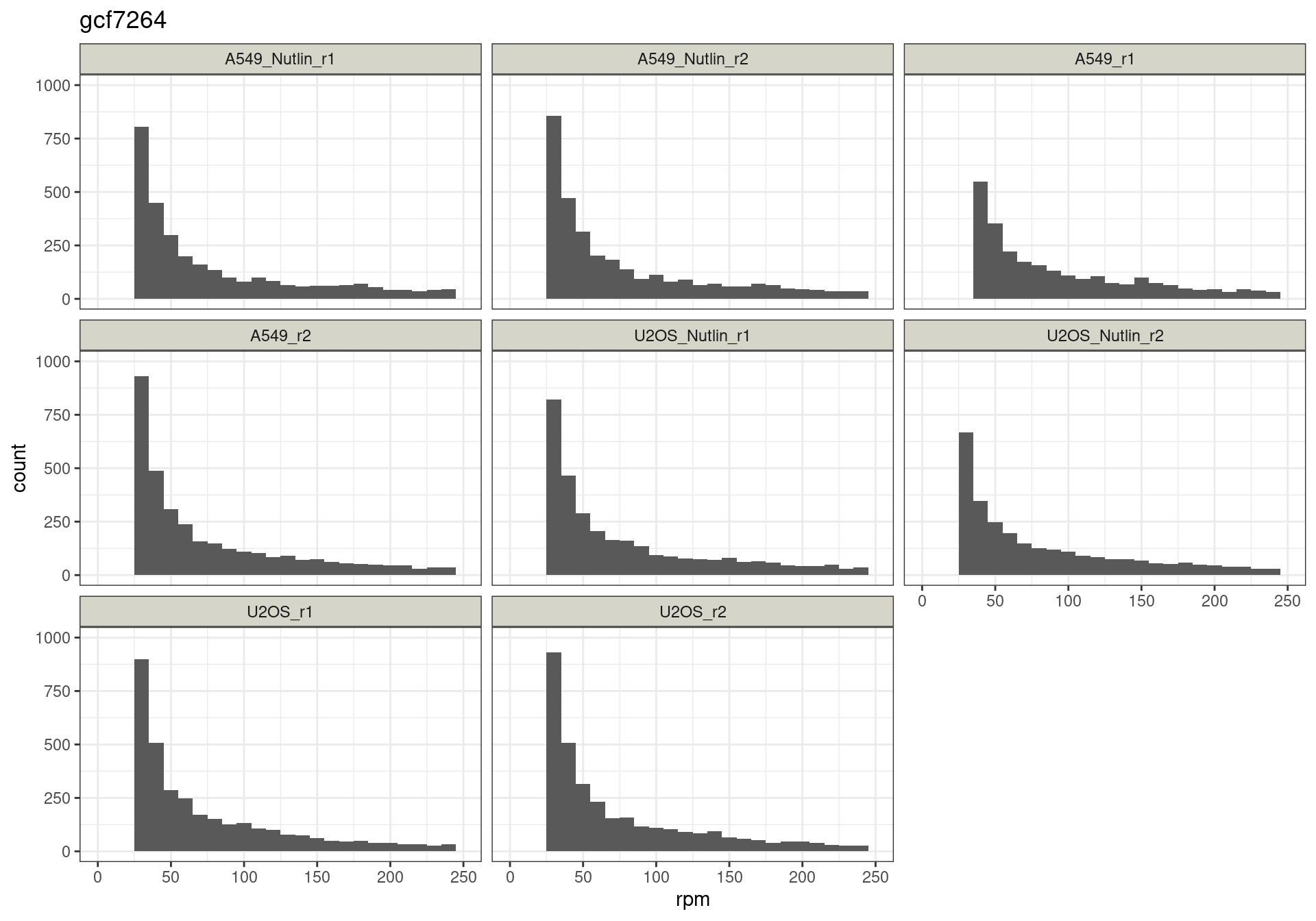
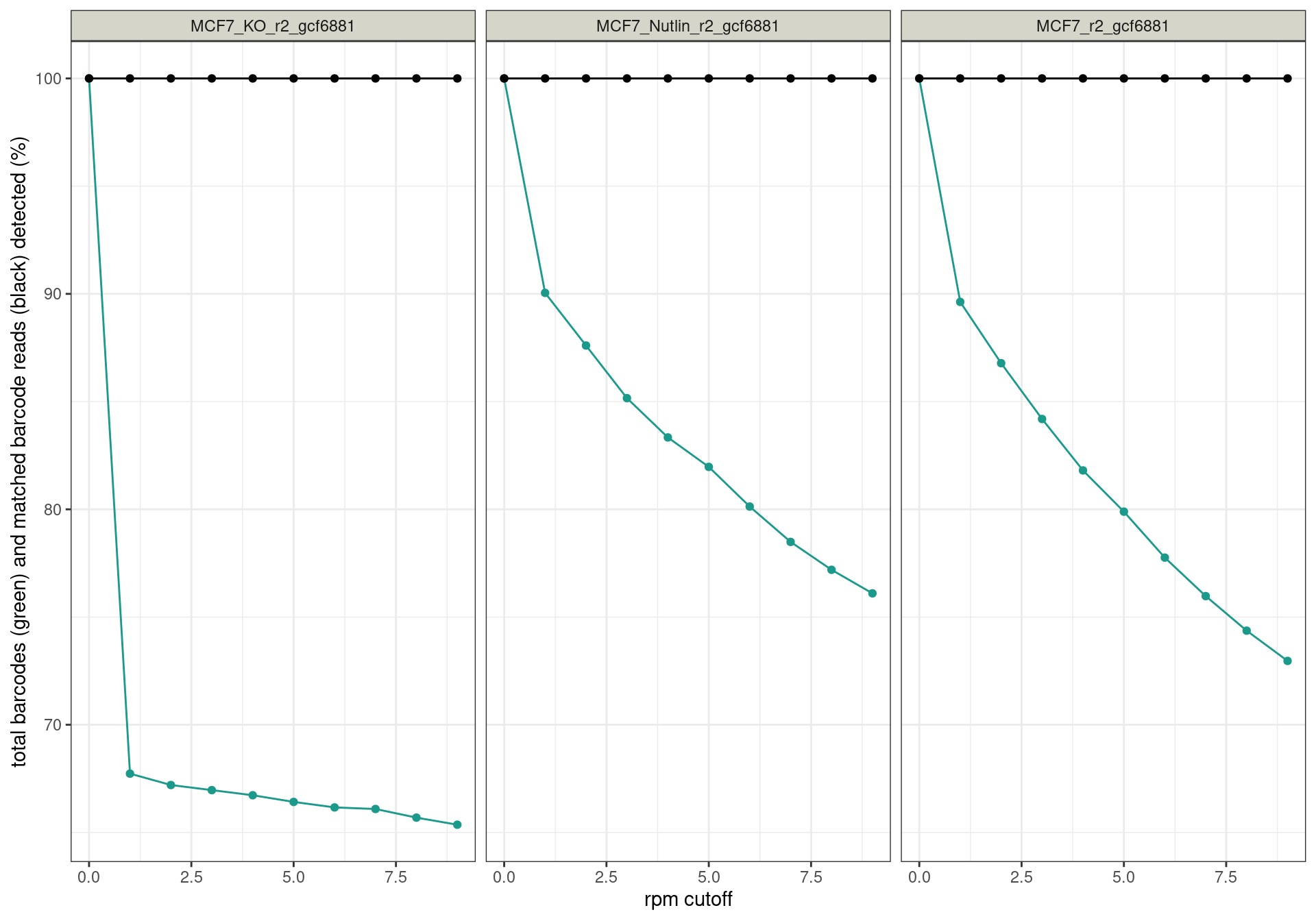 Conclusion: All reads are matched to barcodes that come from the
reporter library. Very good. All barcodes can be found back. At a cutoff
of ~5 rpm, already ~10% of the barcodes are lost. I guess this is to be
expected.
Conclusion: All reads are matched to barcodes that come from the
reporter library. Very good. All barcodes can be found back. At a cutoff
of ~5 rpm, already ~10% of the barcodes are lost. I guess this is to be
expected.
pDNA-cDNA correlation
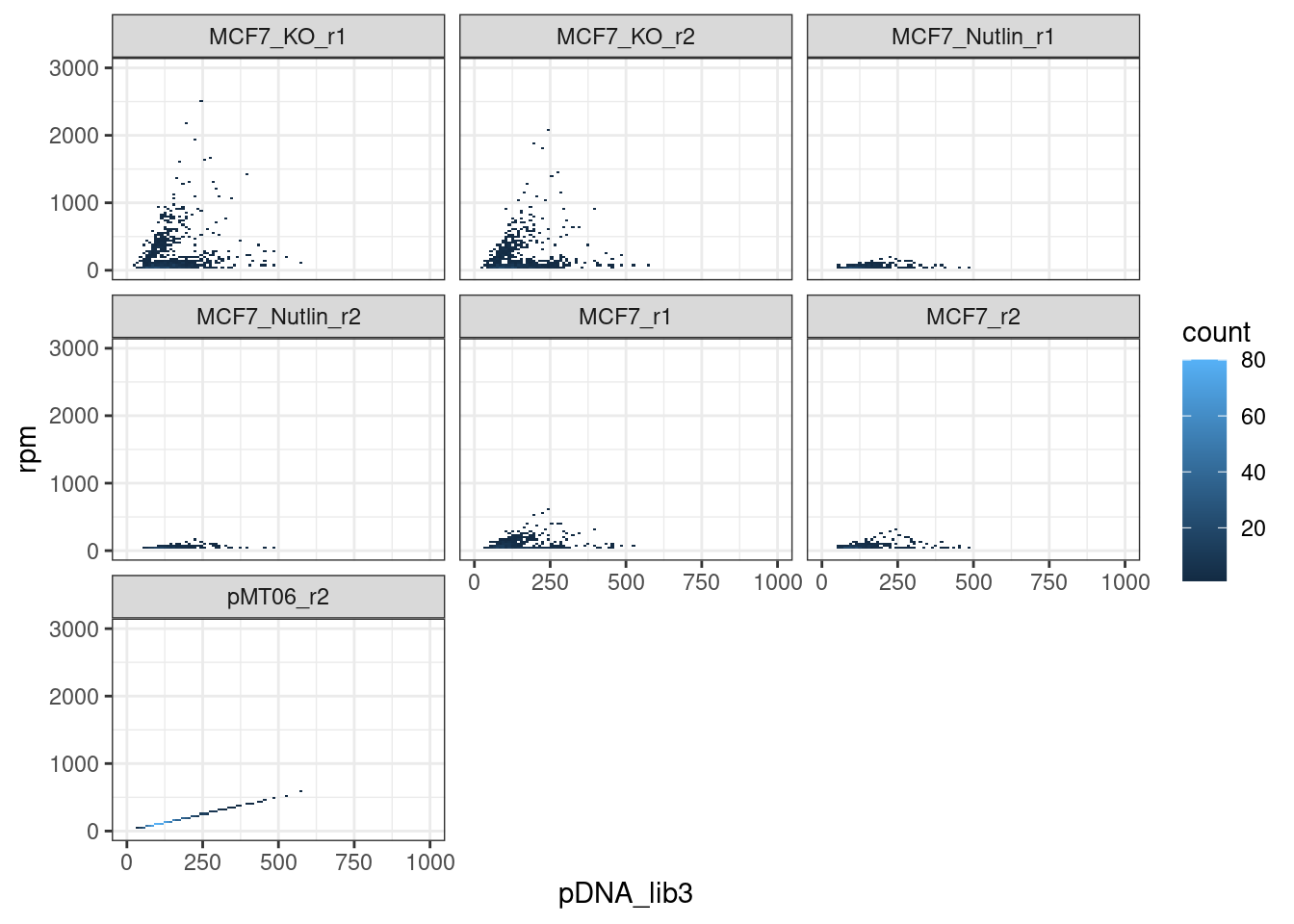
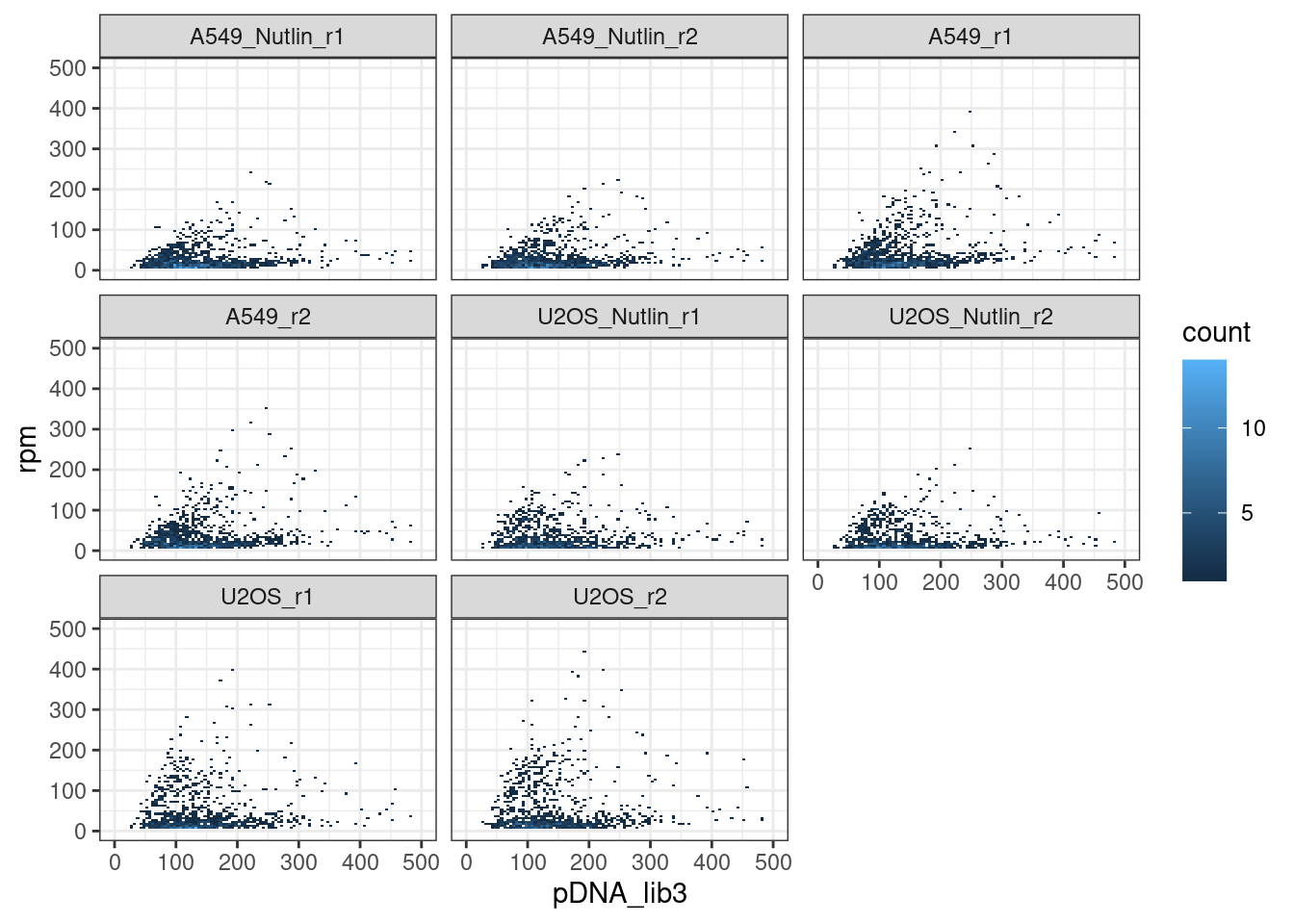
Conclusion: None of the samples correlate with the pDNA input, which means that we are actually measuring the abundance of transcribed barcodes in the cDNA.
Replicate correlation
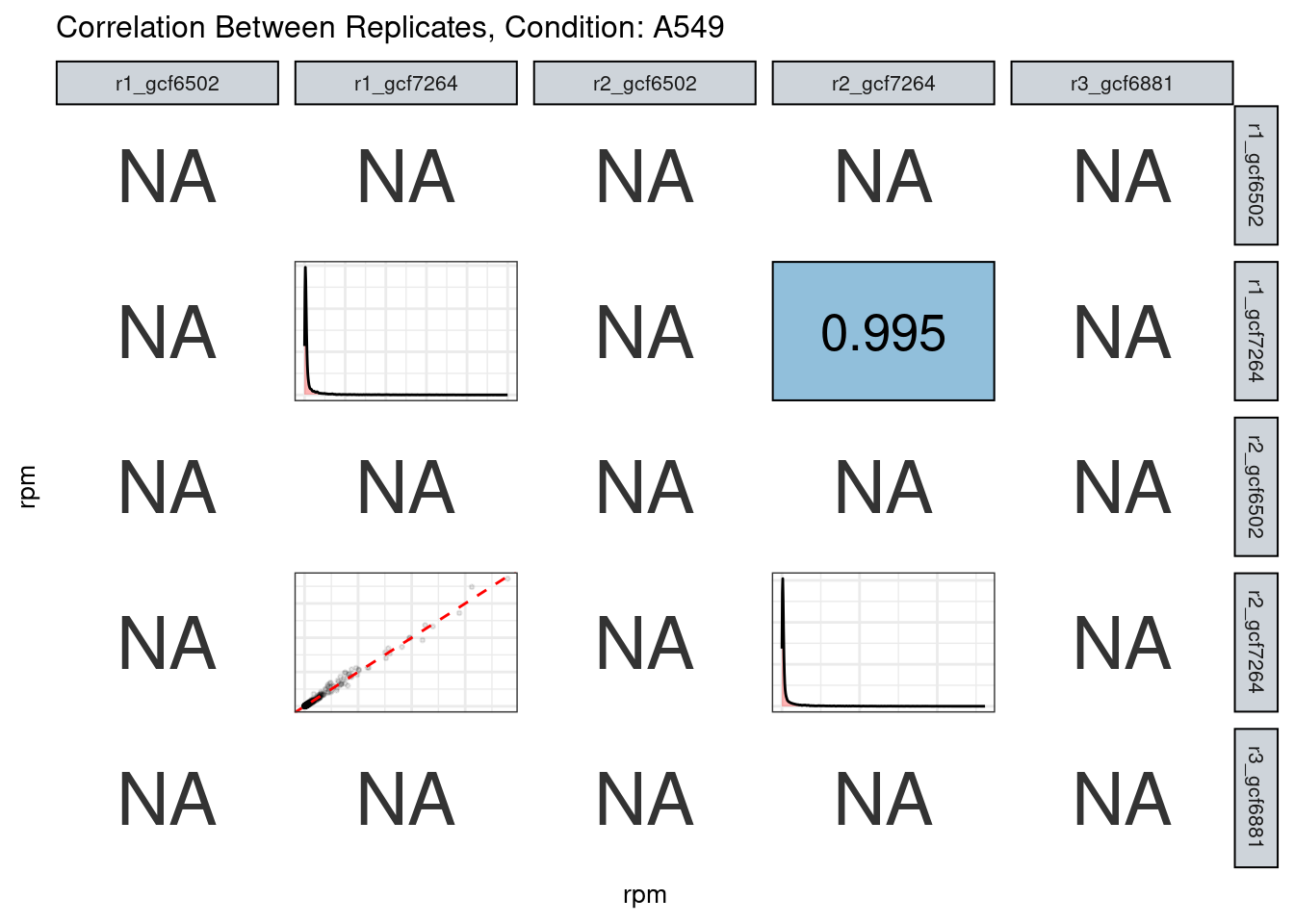
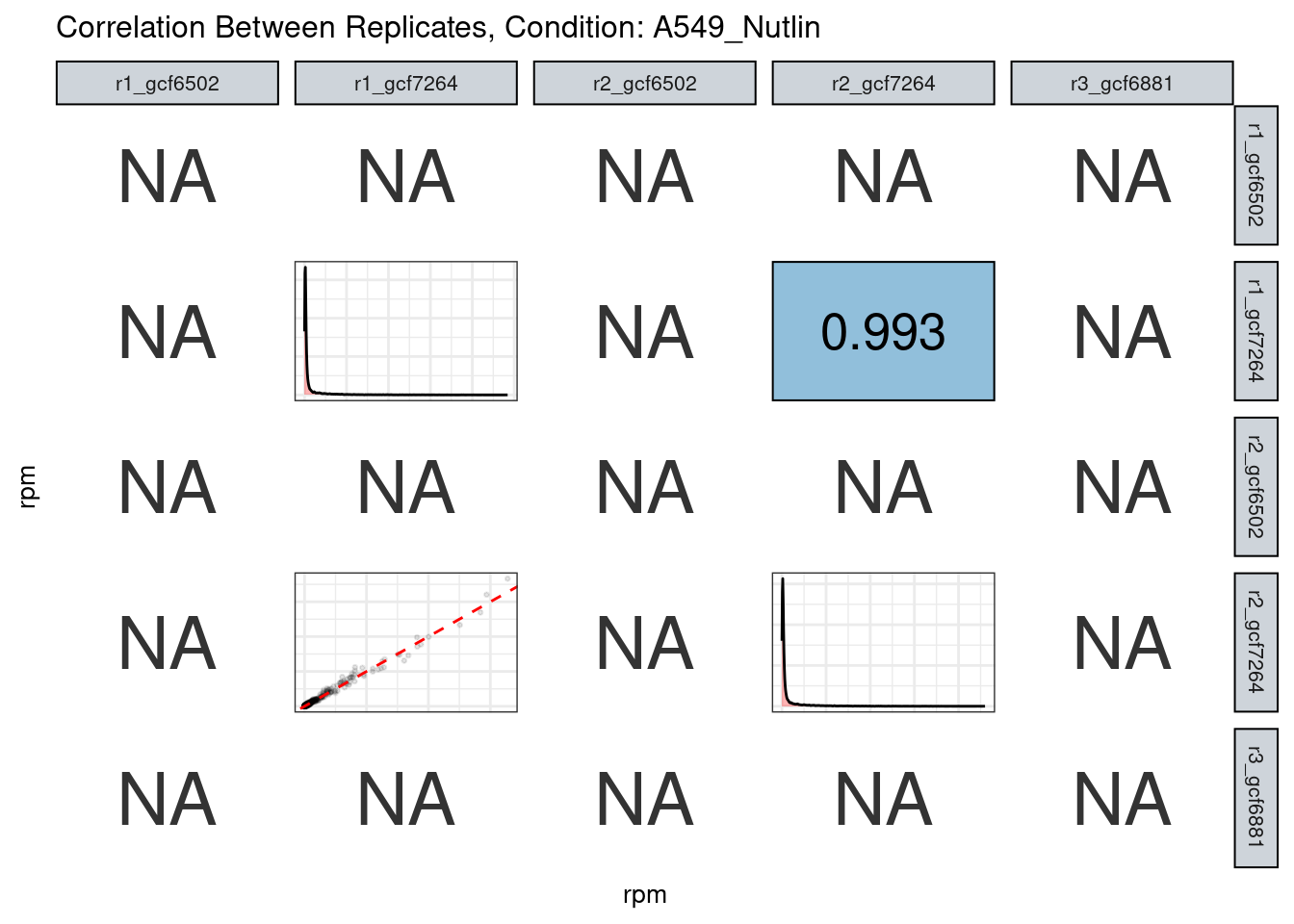
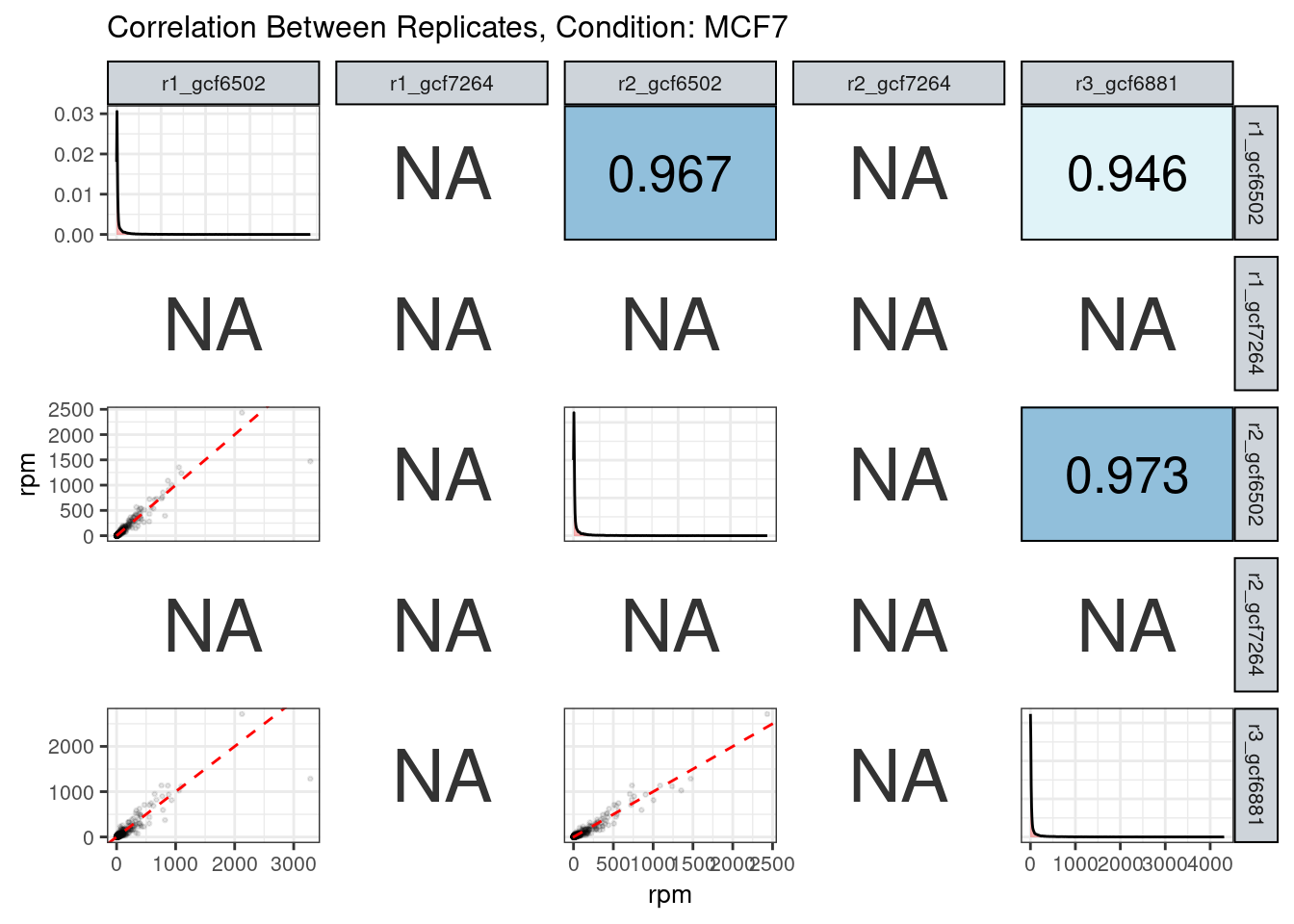
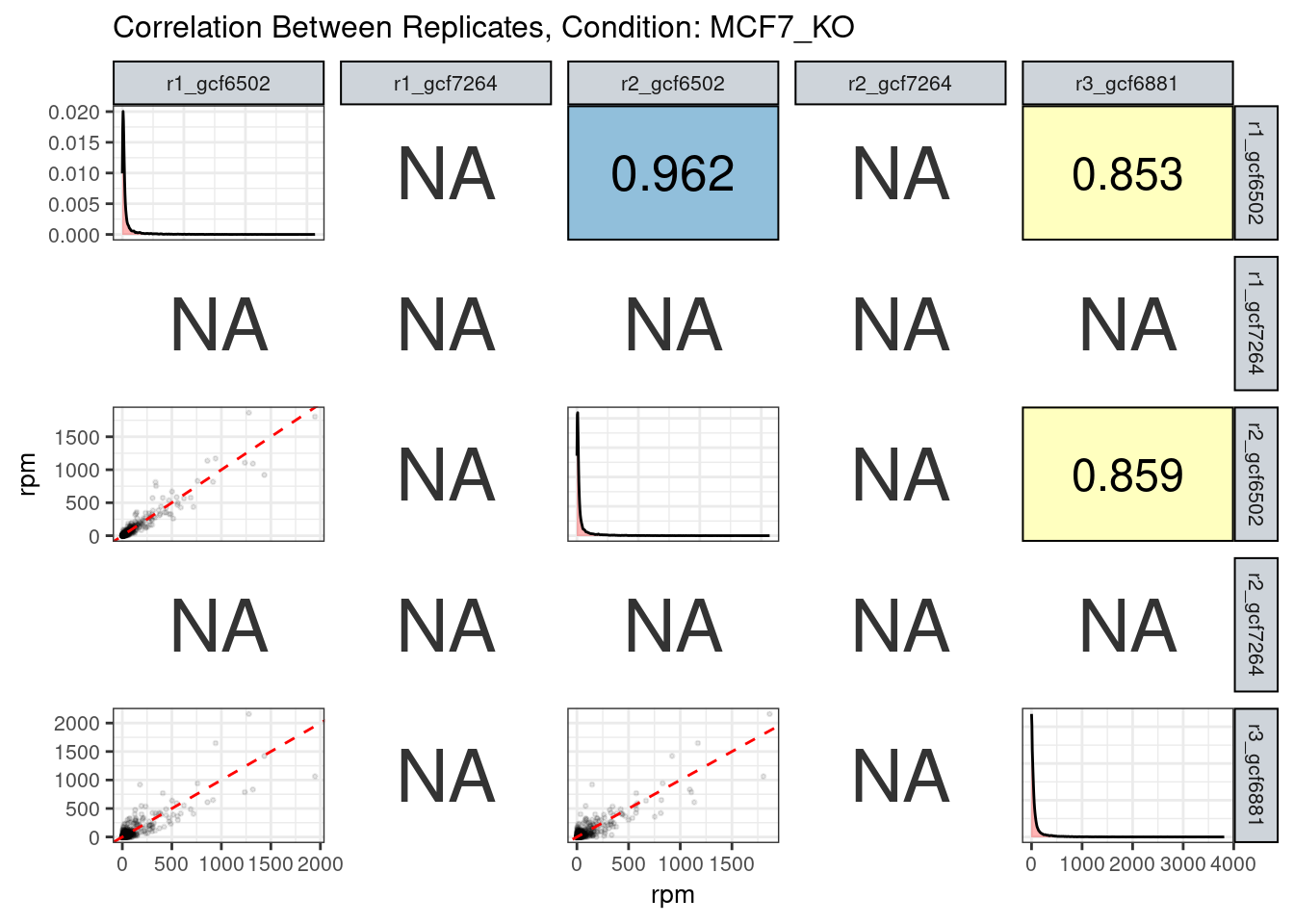
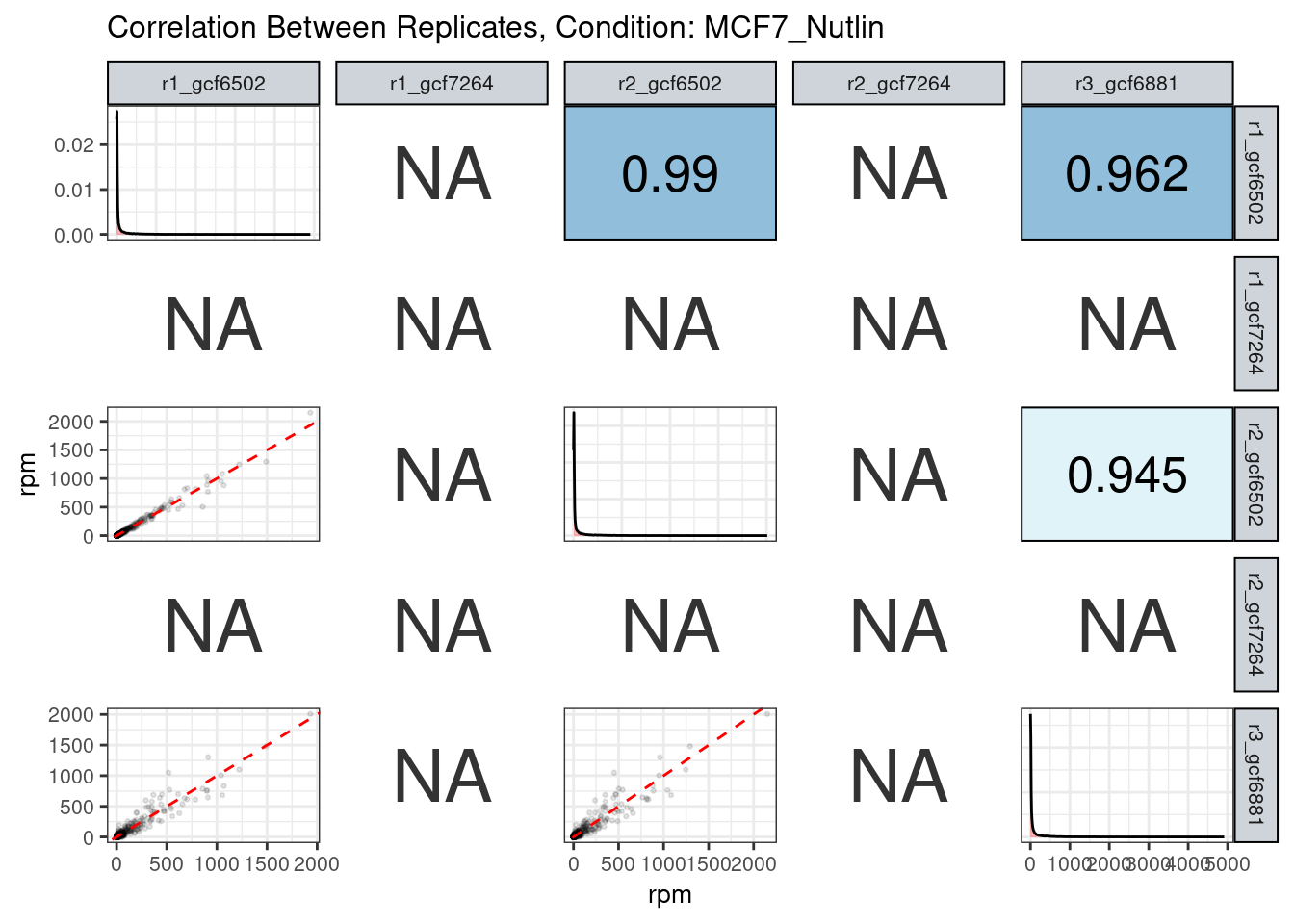
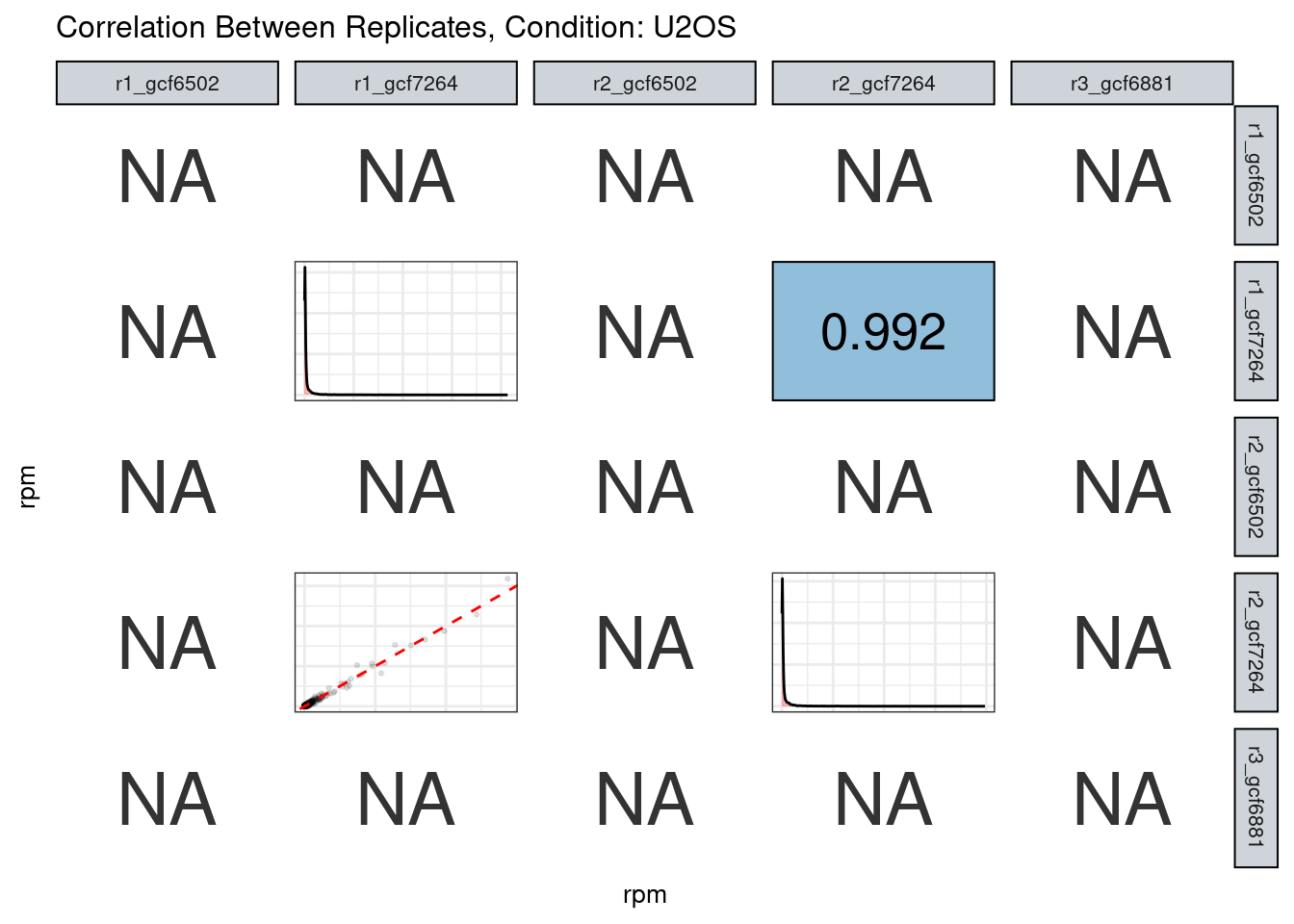
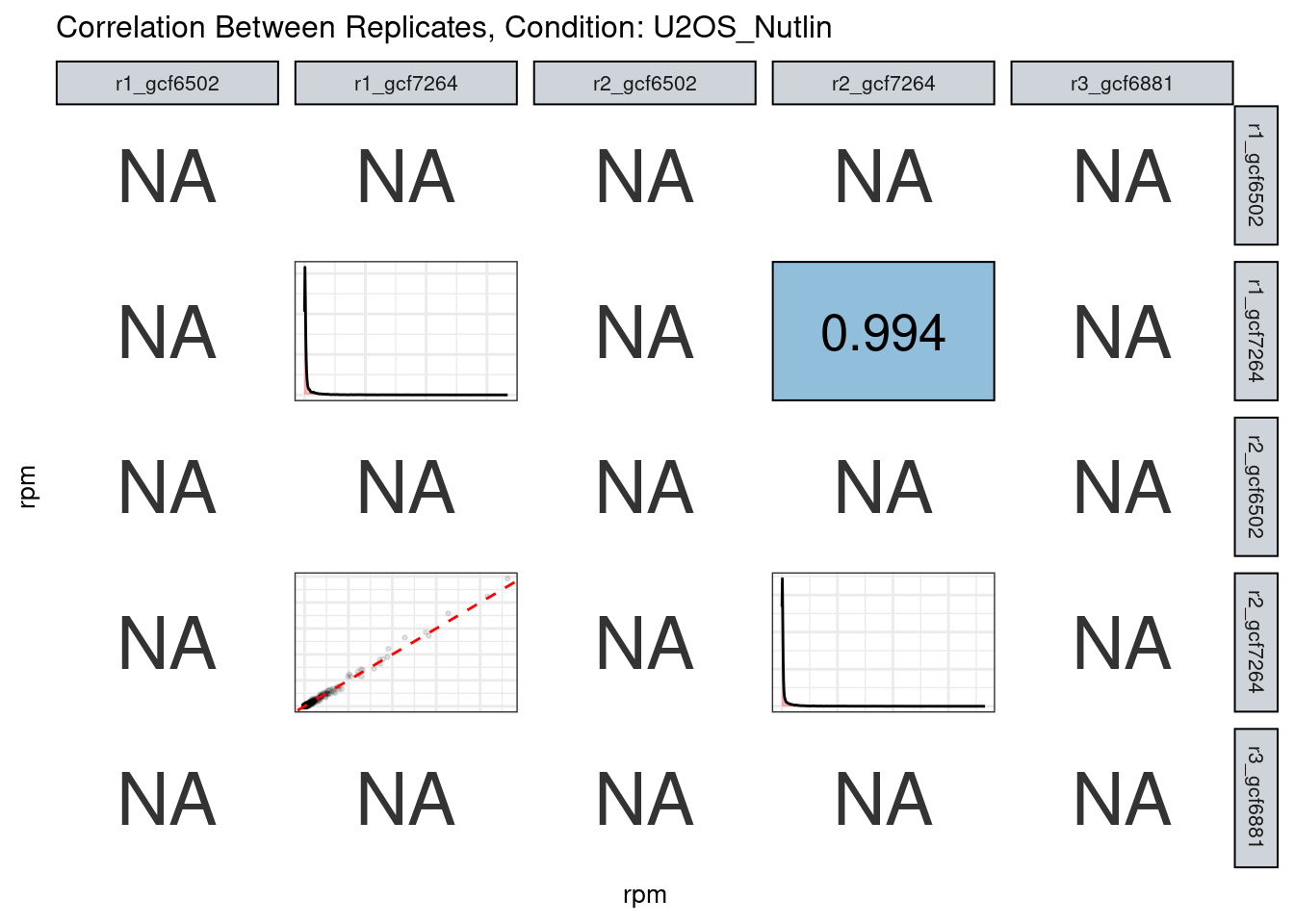 The read counts highly correlate!
The read counts highly correlate!
Annotation of the reporters
Normalization of barcode counts:
Divide cDNA barcode counts through pDNA barcode counts
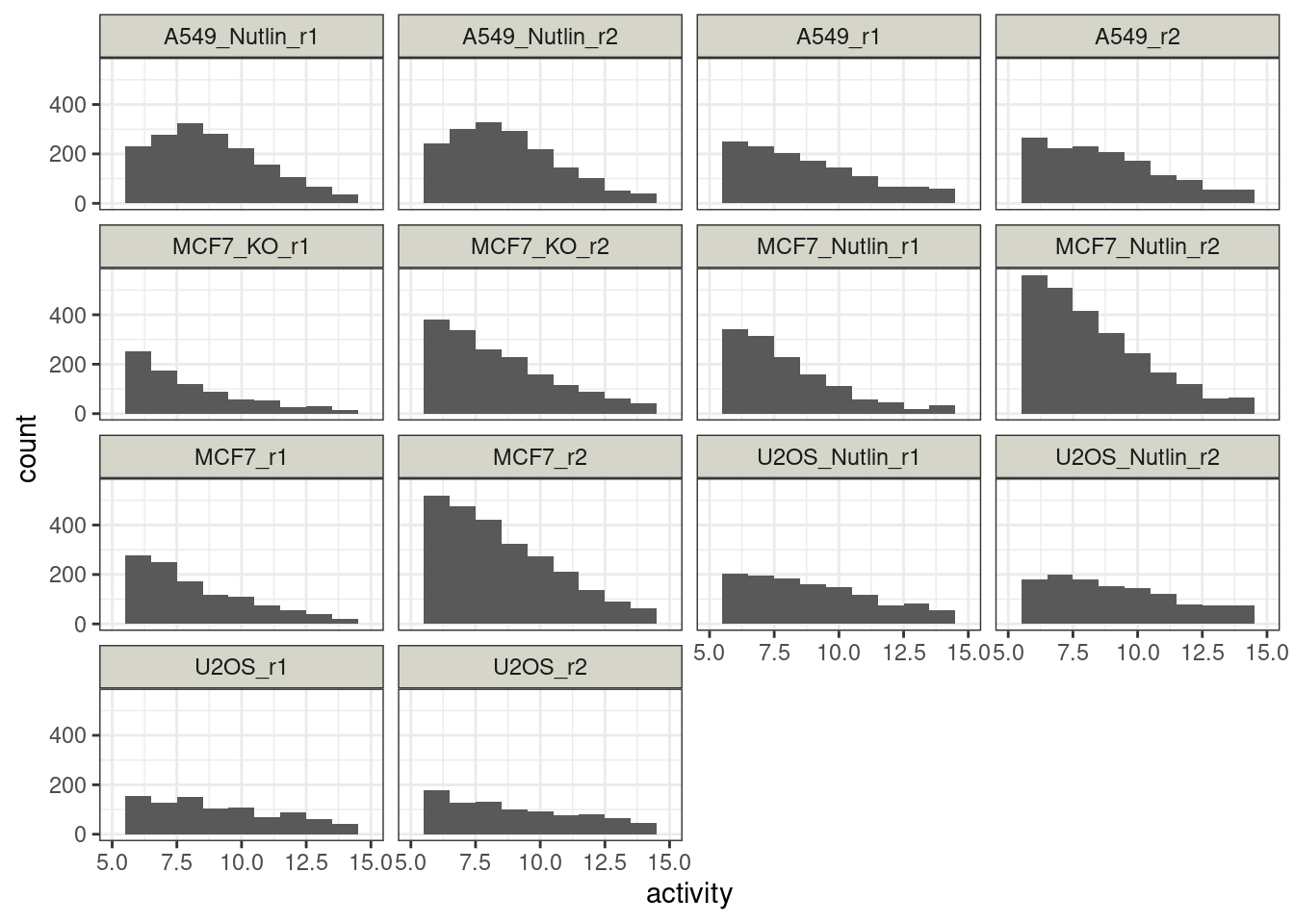
Characterize reporter activities
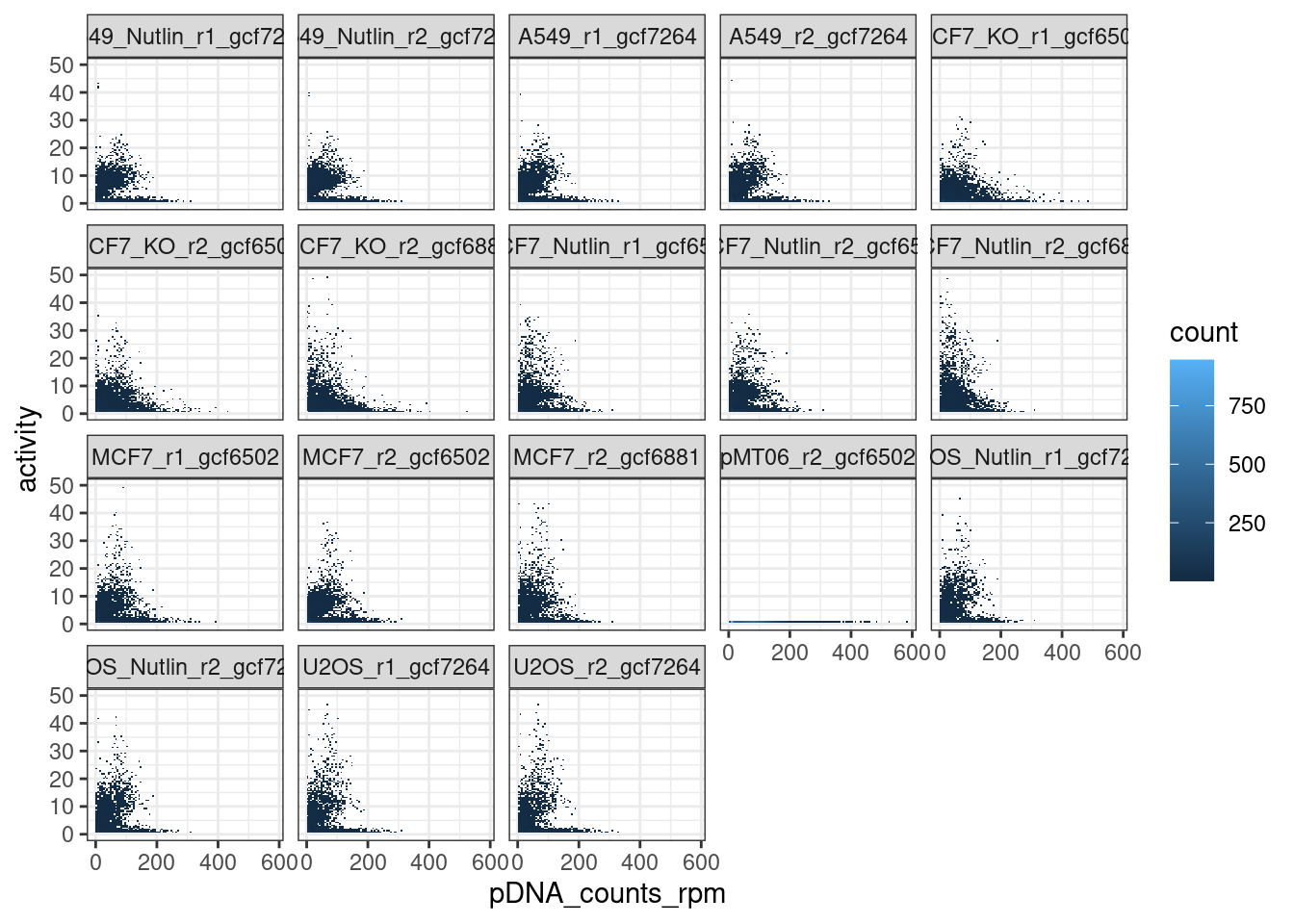
Filtering data
Technical replicate correlations
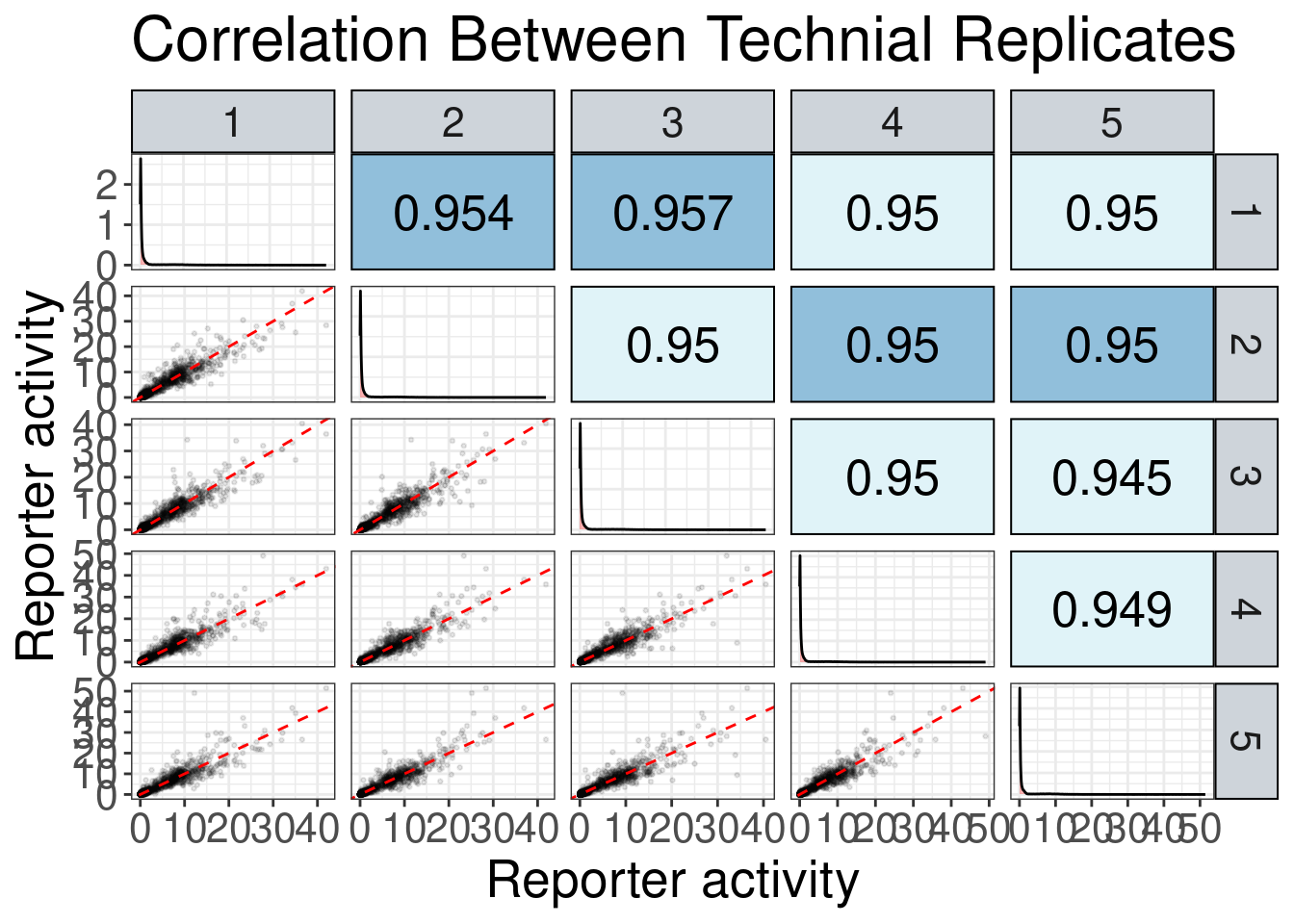 Data correlates very well.
Data correlates very well.
Export data
Session Info
paste("Run time: ",format(Sys.time()-StartTime))## [1] "Run time: 7.879501 mins"getwd()## [1] "/DATA/usr/m.trauernicht/projects/P53_reporter_scan/docs"date()## [1] "Wed Jun 14 09:41:56 2023"sessionInfo()## R version 4.0.5 (2021-03-31)
## Platform: x86_64-pc-linux-gnu (64-bit)
## Running under: Ubuntu 20.04.6 LTS
##
## Matrix products: default
## BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
## LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/liblapack.so.3
##
## locale:
## [1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8 LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8 LC_PAPER=en_US.UTF-8
## [8] LC_NAME=C LC_ADDRESS=C LC_TELEPHONE=C LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
##
## attached base packages:
## [1] stats4 grid parallel stats graphics grDevices utils datasets methods base
##
## other attached packages:
## [1] ggrastr_1.0.1 jtools_2.1.4 glmnetUtils_1.1.8 glmnet_4.1-4 Matrix_1.5-1 randomForest_4.6-14
## [7] ROCR_1.0-11 cowplot_1.1.1 ggforce_0.3.3 maditr_0.8.3 PCAtools_2.2.0 ggrepel_0.9.1
## [13] DESeq2_1.30.1 SummarizedExperiment_1.20.0 Biobase_2.50.0 MatrixGenerics_1.2.1 matrixStats_0.62.0 GenomicRanges_1.42.0
## [19] GenomeInfoDb_1.26.7 IRanges_2.24.1 S4Vectors_0.28.1 BiocGenerics_0.36.1 tidyr_1.2.0 viridis_0.6.2
## [25] viridisLite_0.4.0 ggpointdensity_0.1.0 ggbiplot_0.55 scales_1.2.0 factoextra_1.0.7.999 shiny_1.7.1
## [31] pheatmap_1.0.12 gridExtra_2.3 RColorBrewer_1.1-3 readr_2.1.2 haven_2.5.0 ggbeeswarm_0.6.0
## [37] plotly_4.10.0 tibble_3.1.6 dplyr_1.0.8 vwr_0.3.0 latticeExtra_0.6-29 lattice_0.20-41
## [43] stringdist_0.9.8 GGally_2.1.2 ggpubr_0.4.0 ggplot2_3.4.0 stringr_1.4.0 plyr_1.8.7
## [49] data.table_1.14.2
##
## loaded via a namespace (and not attached):
## [1] backports_1.4.1 lazyeval_0.2.2 splines_4.0.5 crosstalk_1.2.0 BiocParallel_1.24.1 digest_0.6.29 foreach_1.5.2
## [8] htmltools_0.5.2 fansi_1.0.3 magrittr_2.0.3 memoise_2.0.1 tzdb_0.3.0 annotate_1.68.0 vroom_1.5.7
## [15] prettyunits_1.1.1 jpeg_0.1-9 colorspace_2.0-3 blob_1.2.3 gitcreds_0.1.1 xfun_0.30 crayon_1.5.1
## [22] RCurl_1.98-1.6 jsonlite_1.8.0 genefilter_1.72.1 iterators_1.0.14 survival_3.2-10 glue_1.6.2 polyclip_1.10-0
## [29] gtable_0.3.0 zlibbioc_1.36.0 XVector_0.30.0 DelayedArray_0.16.3 car_3.0-12 BiocSingular_1.6.0 shape_1.4.6
## [36] abind_1.4-5 DBI_1.1.2 rstatix_0.7.0 Rcpp_1.0.8.3 progress_1.2.2 xtable_1.8-4 dqrng_0.3.0
## [43] bit_4.0.4 rsvd_1.0.5 htmlwidgets_1.5.4 httr_1.4.2 ellipsis_0.3.2 farver_2.1.0 pkgconfig_2.0.3
## [50] reshape_0.8.9 XML_3.99-0.9 sass_0.4.1 locfit_1.5-9.4 utf8_1.2.2 labeling_0.4.2 tidyselect_1.1.2
## [57] rlang_1.0.6 reshape2_1.4.4 later_1.3.0 AnnotationDbi_1.52.0 munsell_0.5.0 tools_4.0.5 cachem_1.0.6
## [64] cli_3.4.1 generics_0.1.2 RSQLite_2.2.12 broom_0.8.0 evaluate_0.15 fastmap_1.1.0 yaml_2.3.5
## [71] knitr_1.38 bit64_4.0.5 pander_0.6.5 purrr_0.3.4 sparseMatrixStats_1.2.1 mime_0.12 compiler_4.0.5
## [78] rstudioapi_0.13 beeswarm_0.4.0 png_0.1-7 ggsignif_0.6.3 tweenr_1.0.2 geneplotter_1.68.0 bslib_0.3.1
## [85] stringi_1.7.6 highr_0.9 forcats_0.5.1 vctrs_0.5.1 pillar_1.7.0 lifecycle_1.0.3 jquerylib_0.1.4
## [92] bitops_1.0-7 irlba_2.3.5 httpuv_1.6.5 R6_2.5.1 promises_1.2.0.1 vipor_0.4.5 codetools_0.2-18
## [99] MASS_7.3-53.1 assertthat_0.2.1 withr_2.5.0 GenomeInfoDbData_1.2.4 hms_1.1.1 beachmat_2.6.4 rmarkdown_2.13
## [106] DelayedMatrixStats_1.12.3 carData_3.0-5