TP53 reporter library scan - detailed analysis
Max Trauernicht
Netherlands Cancer Institute - van Steensel labm.trauernicht@nki.nl
14/06/2023
Introduction
~6,000 barcoded TP53 reporters were probed in MCF7 TP53WT/KO cells and stimulated with Nutlin-3a. I previously processed the raw sequencing data, quantified the pDNA data and normalized the cDNA data. In this script, a detailed dissection of the reporter activities will be carried out to understand how TP53 drives transcription and to identify the most sensitive TP53 reporters.
Setup
Libraries
Functions
Load data
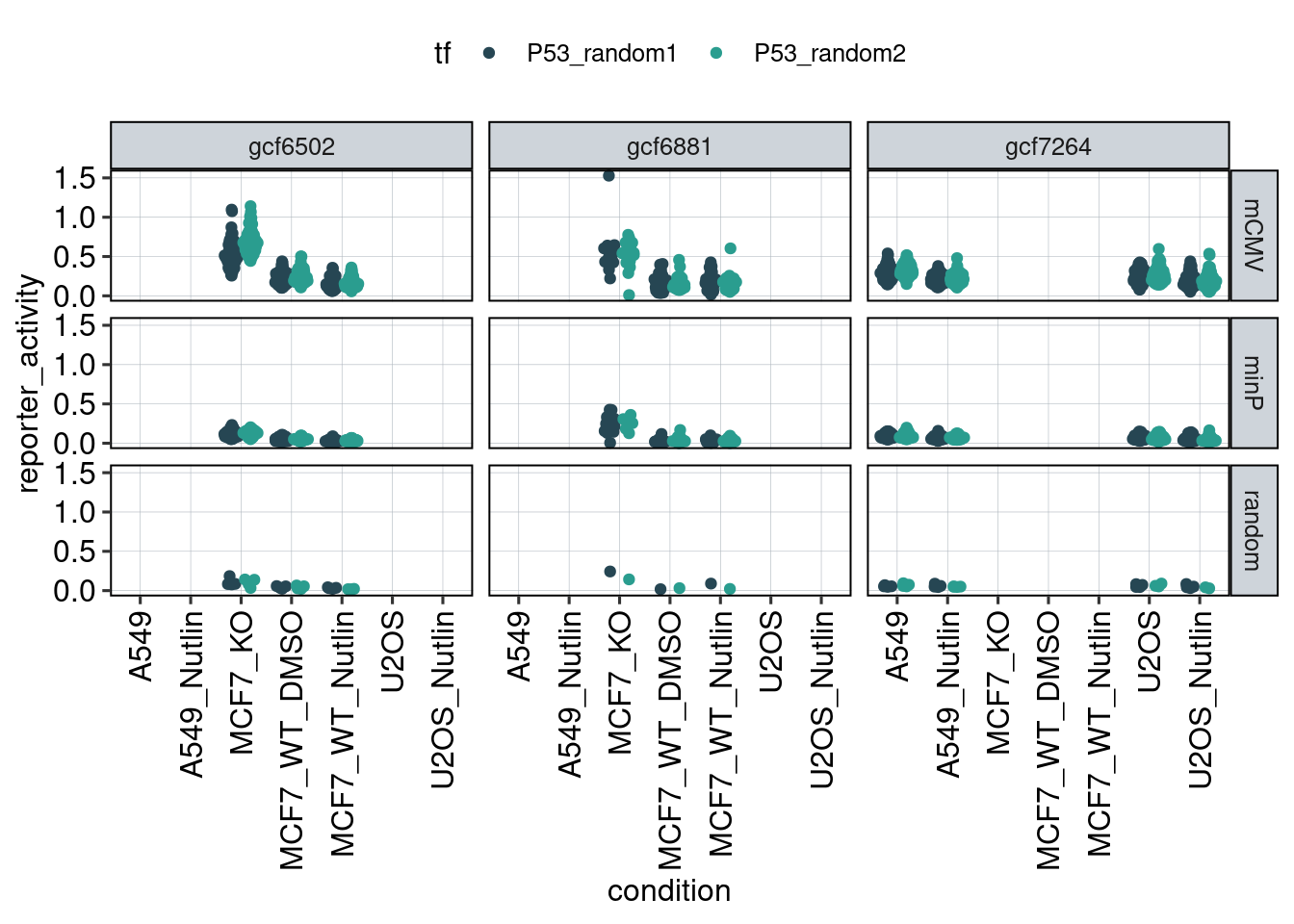
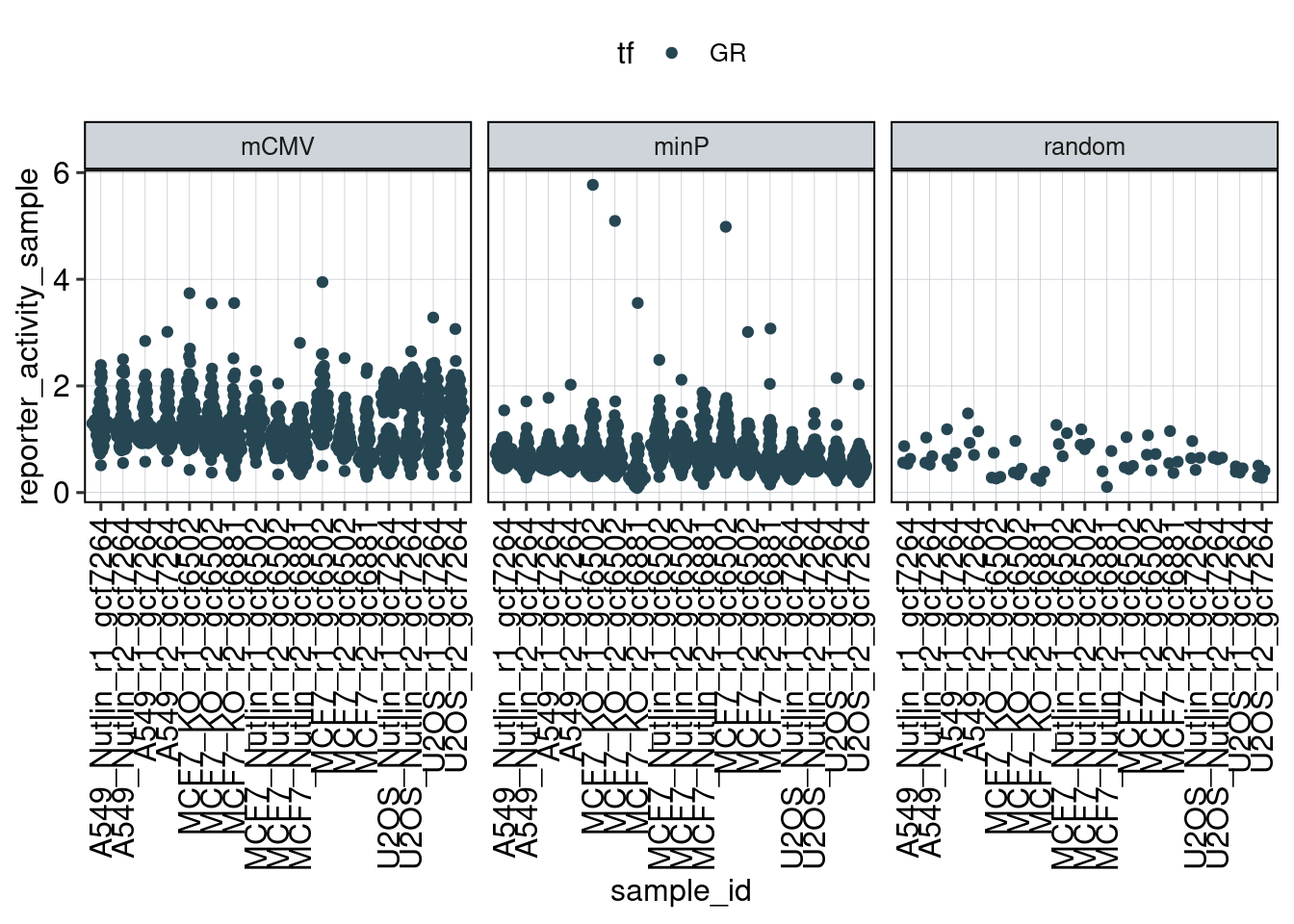
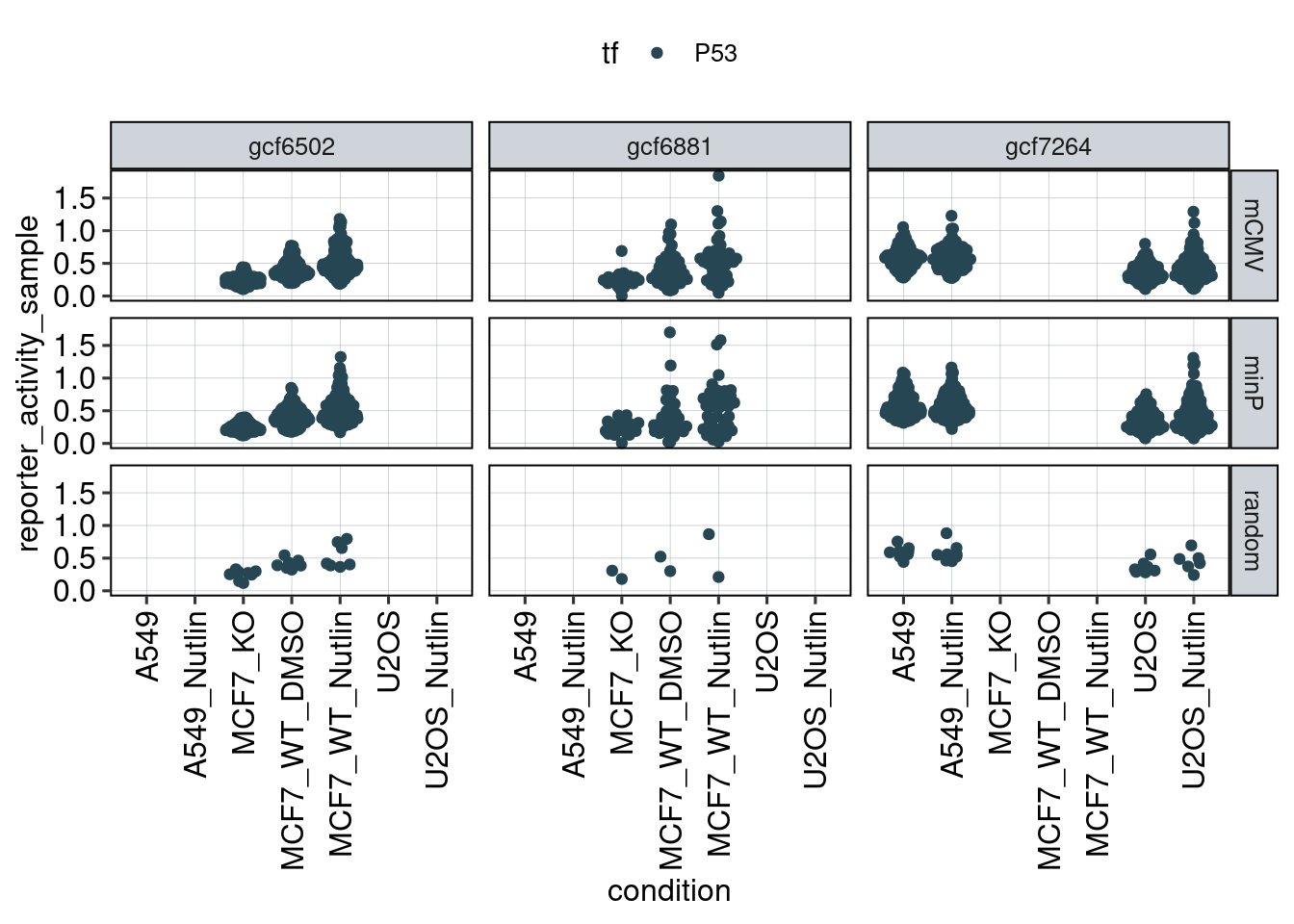
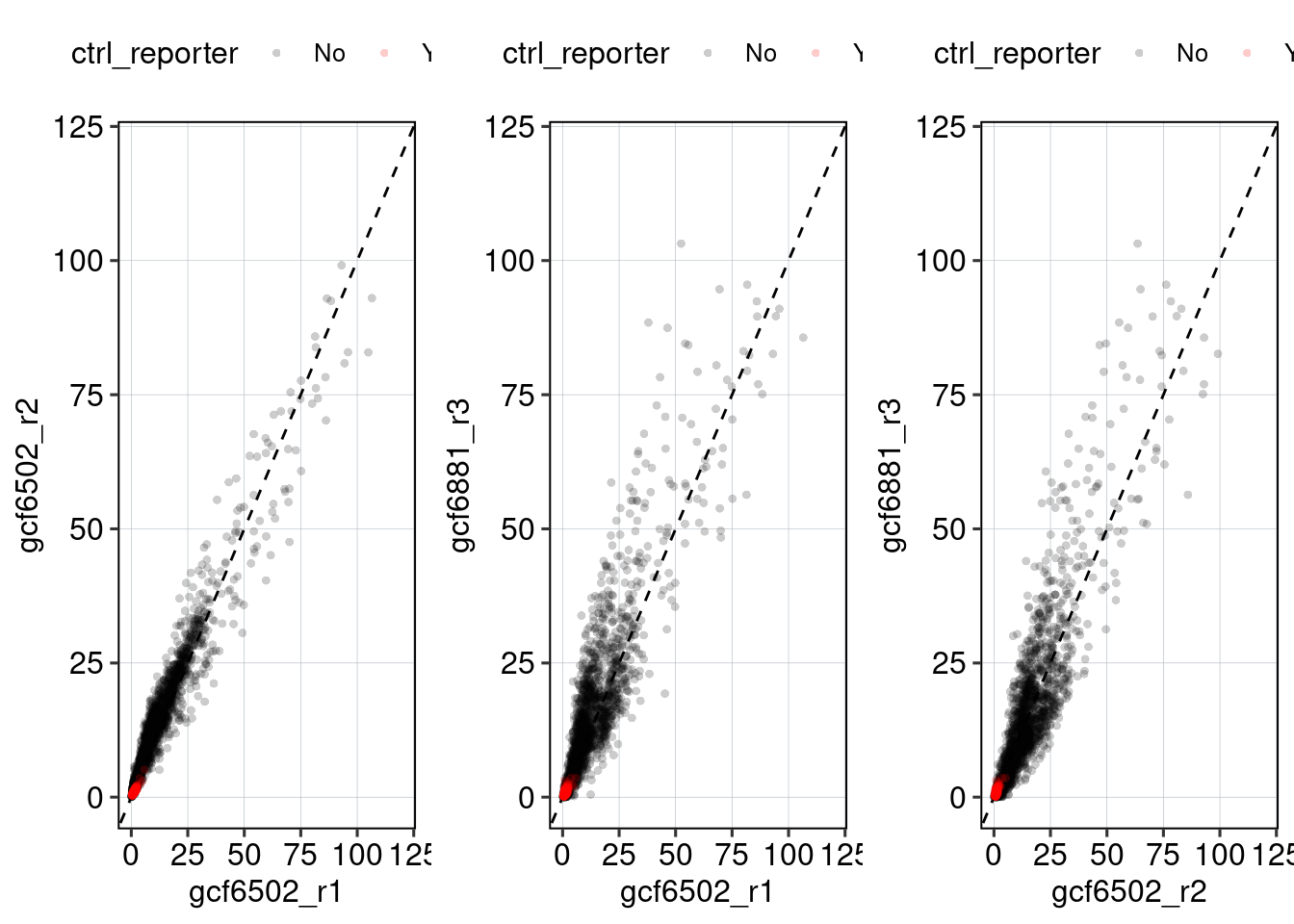
Figure 1: Characterize P53 activities per condition
Aim: I want to characterize the reporter activity distributions in
the tested conditions. Does Nutlin boost P53 reporter activity and is
P53 inactive in the KO cells?
## [1] 0.9685877## [1] 0.9036356## [1] 0.903232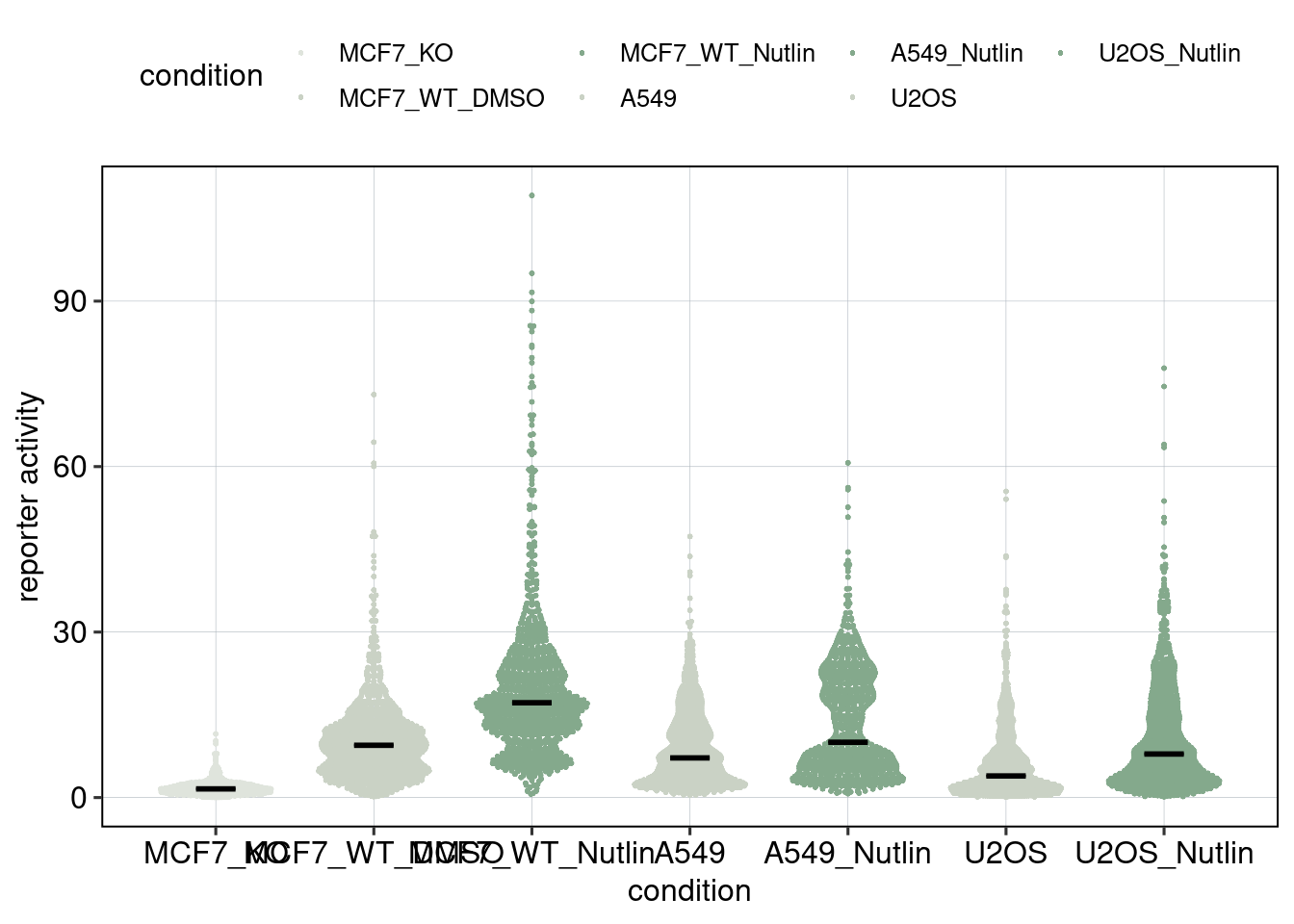
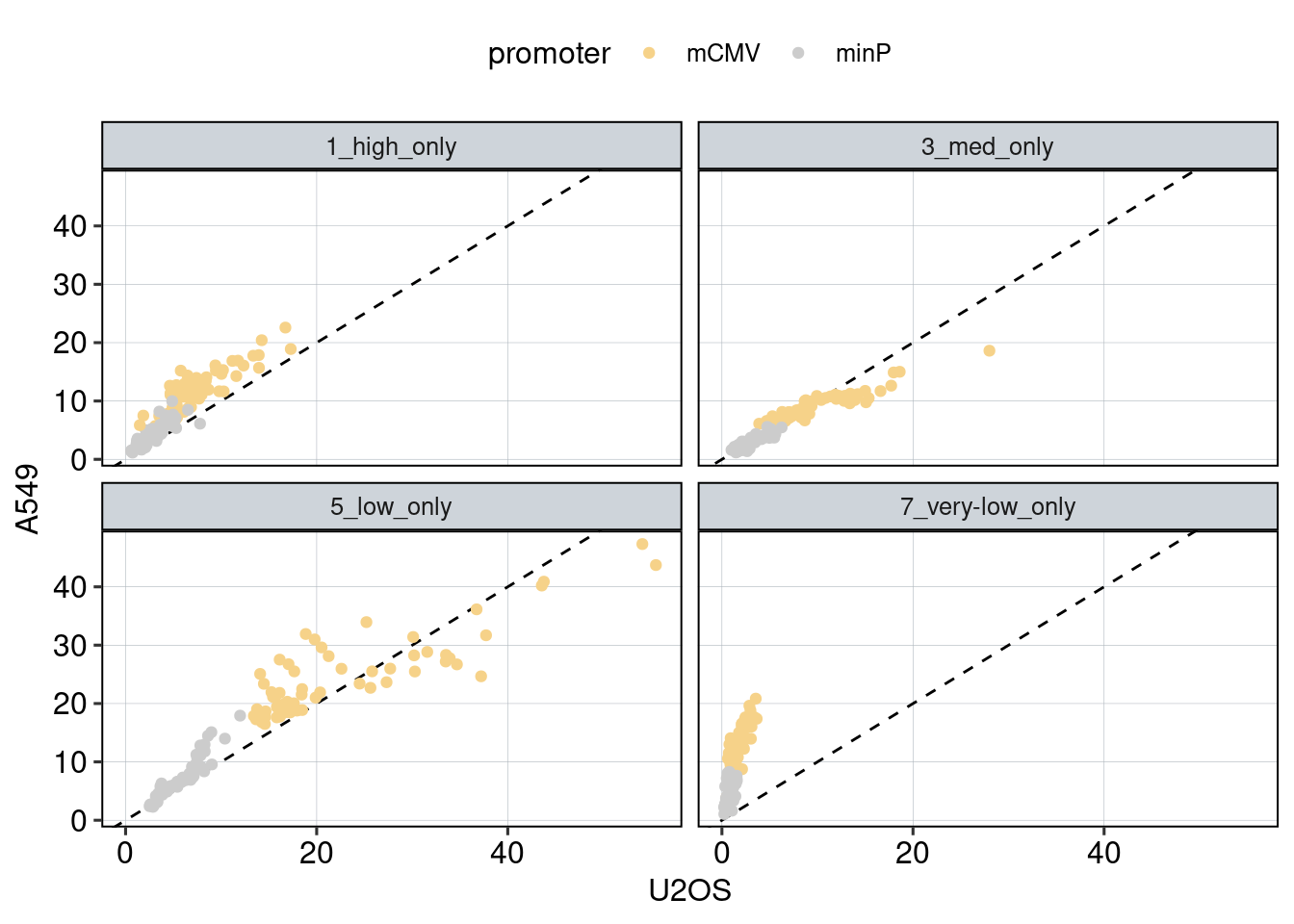
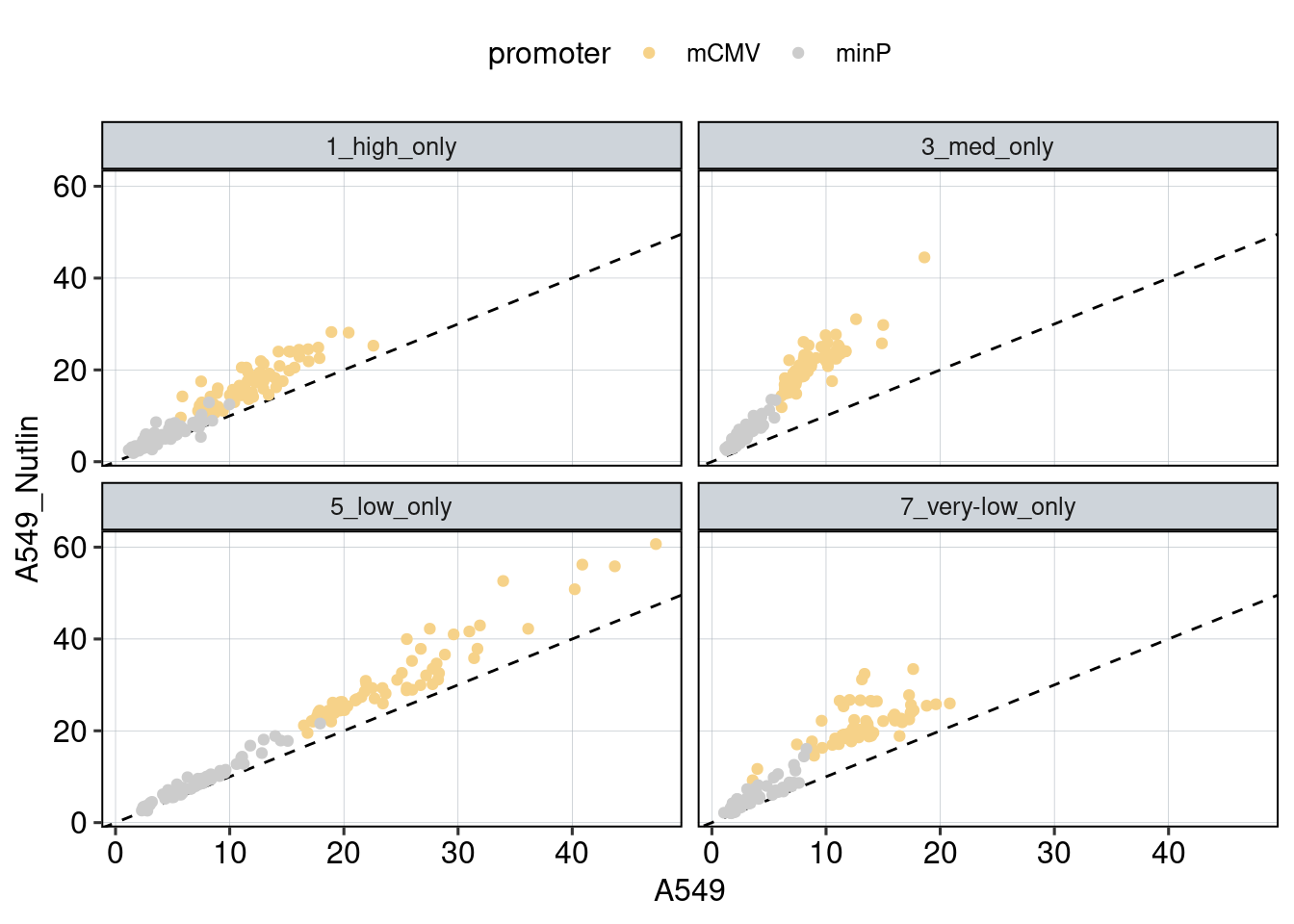
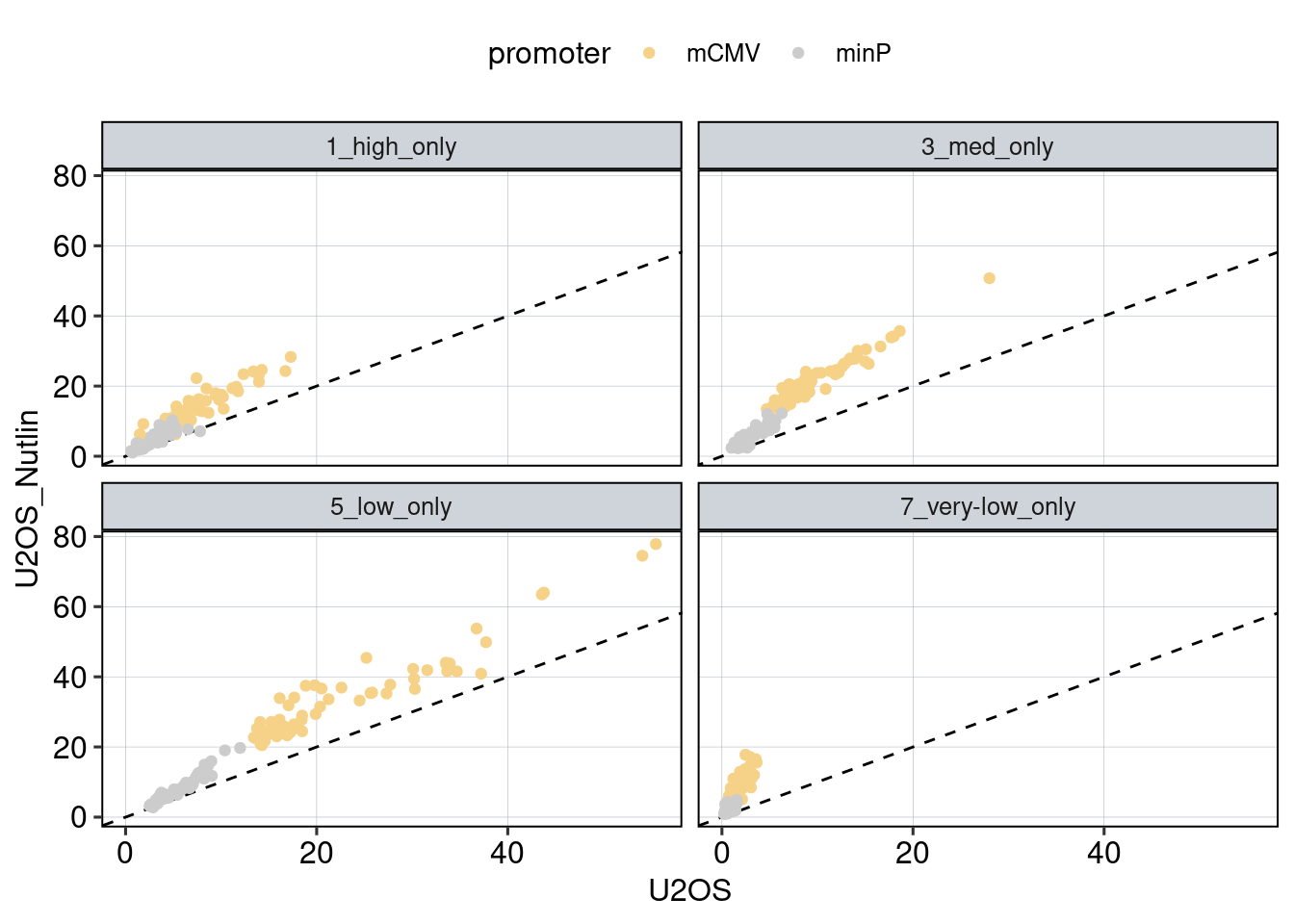
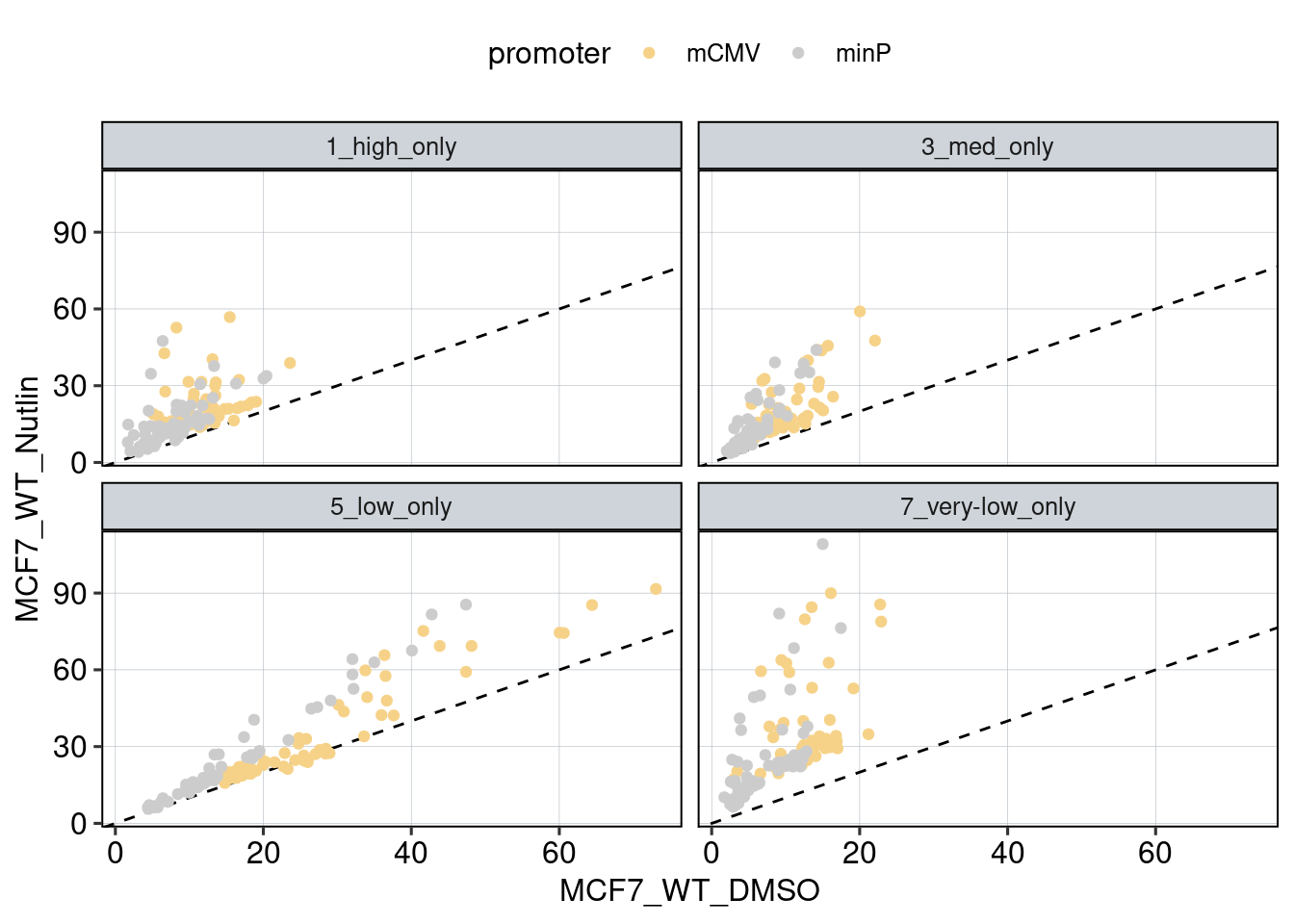
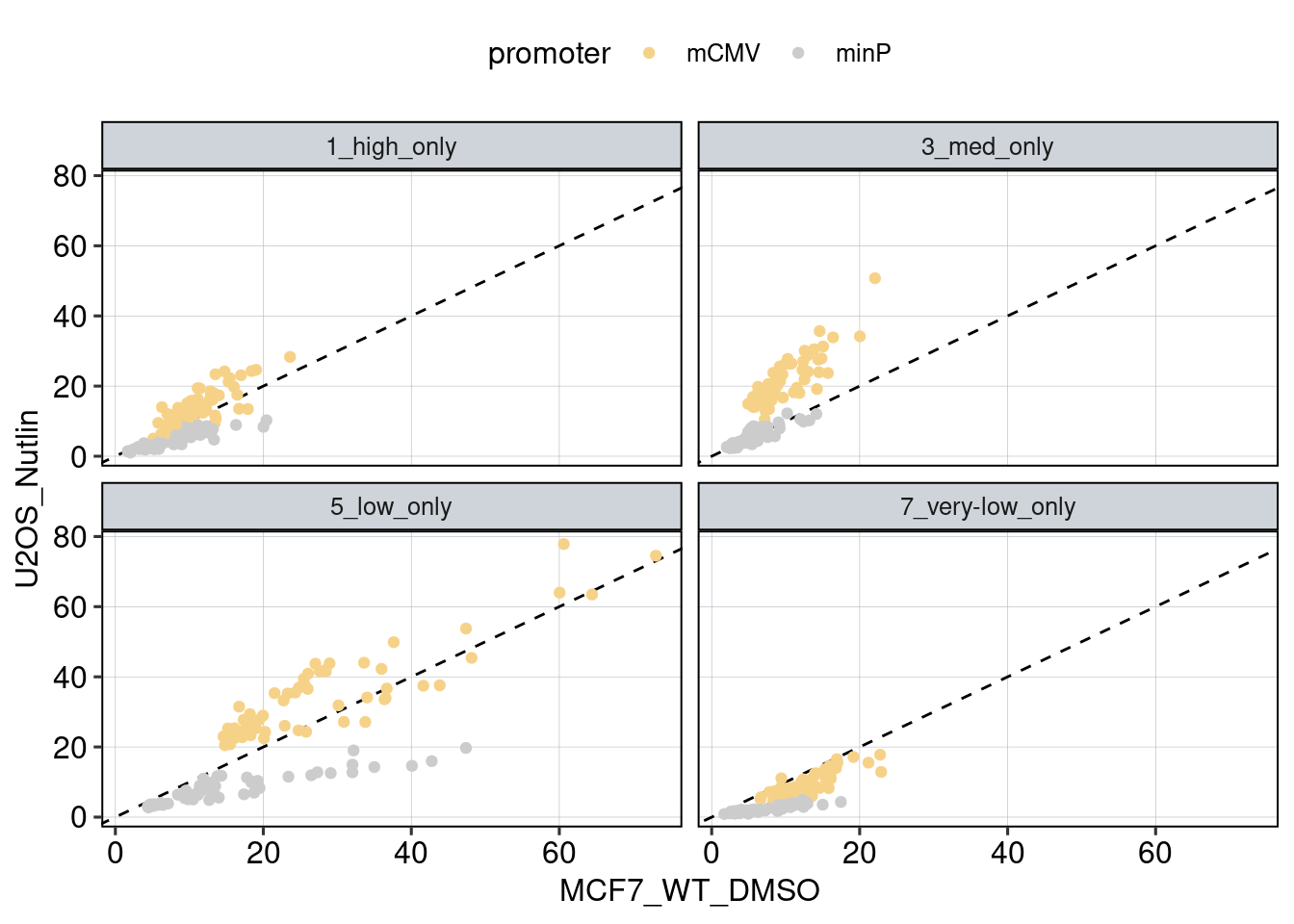
Conclusion: 1F: Replicates do correlate well. 1G: Negative controls are inactive compared to P53 reporters. P53 reporters become more active in WT cells and even more active upon Nutlin stimulation.
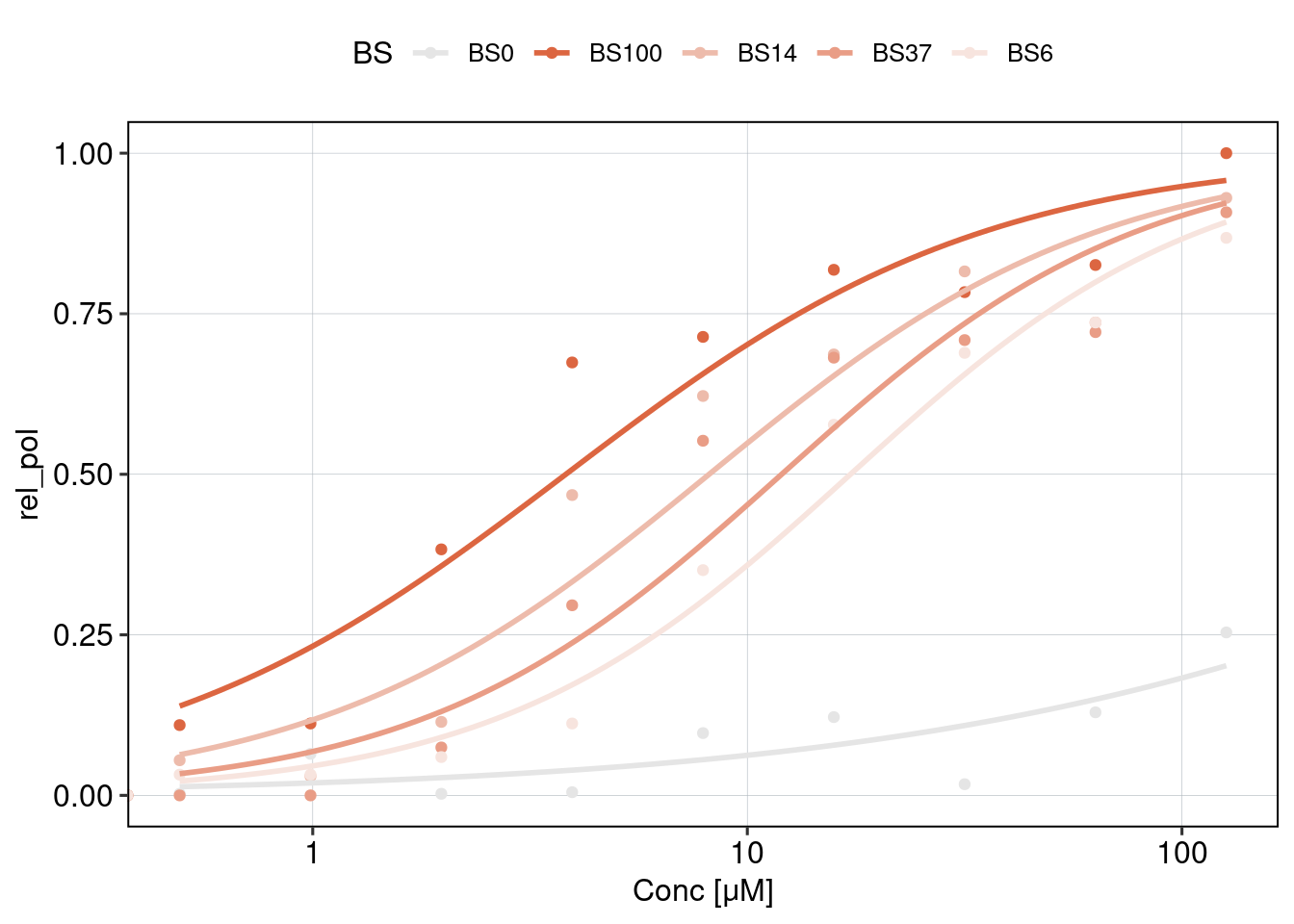
Figure 2: Effect of affinity and binding sites + binding site positioning
Aim: How does the binding site affinity, copy number, and their respective positioning affect reporter activity?
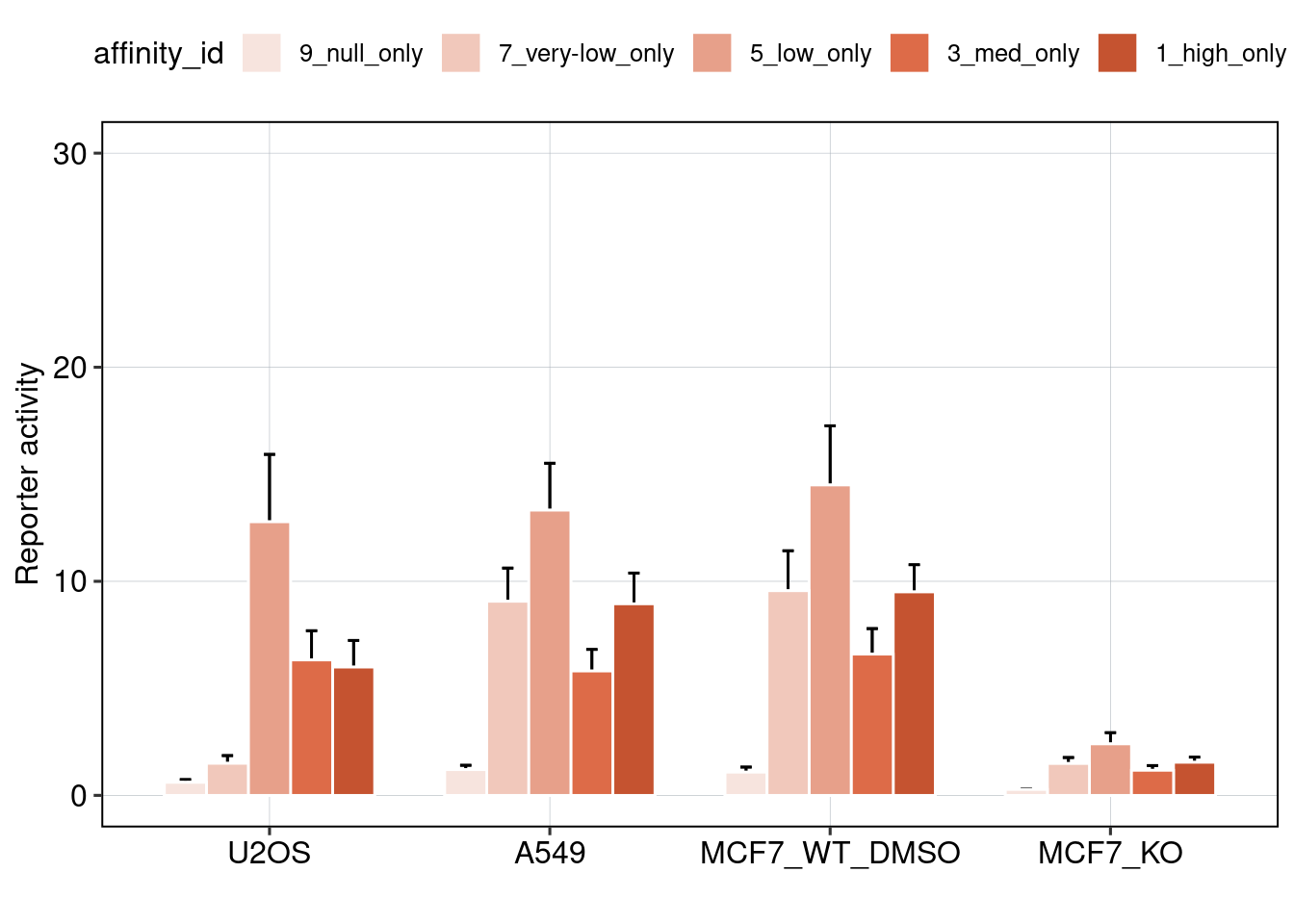
## [1] 0.006910845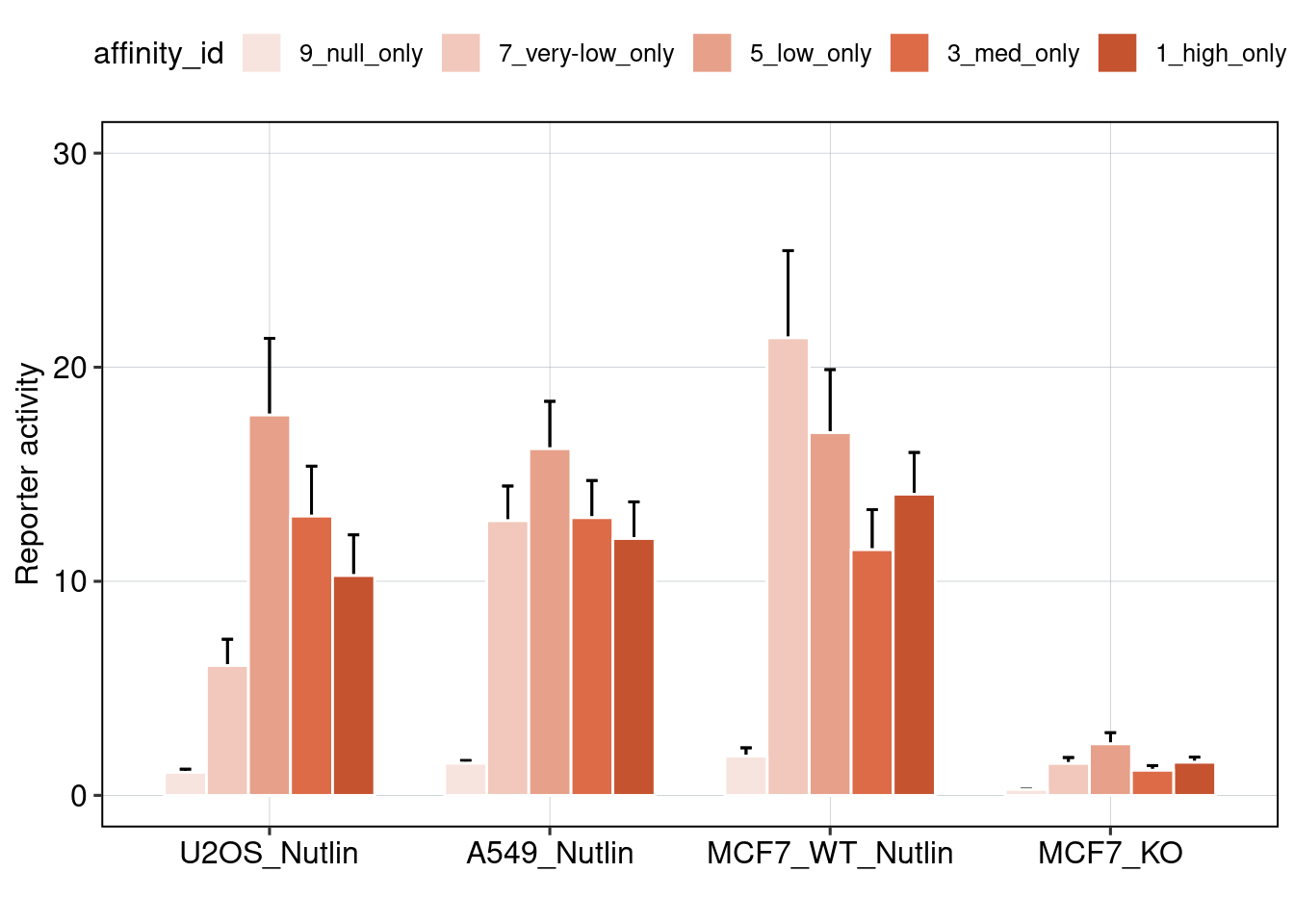
## [1] 0.02978148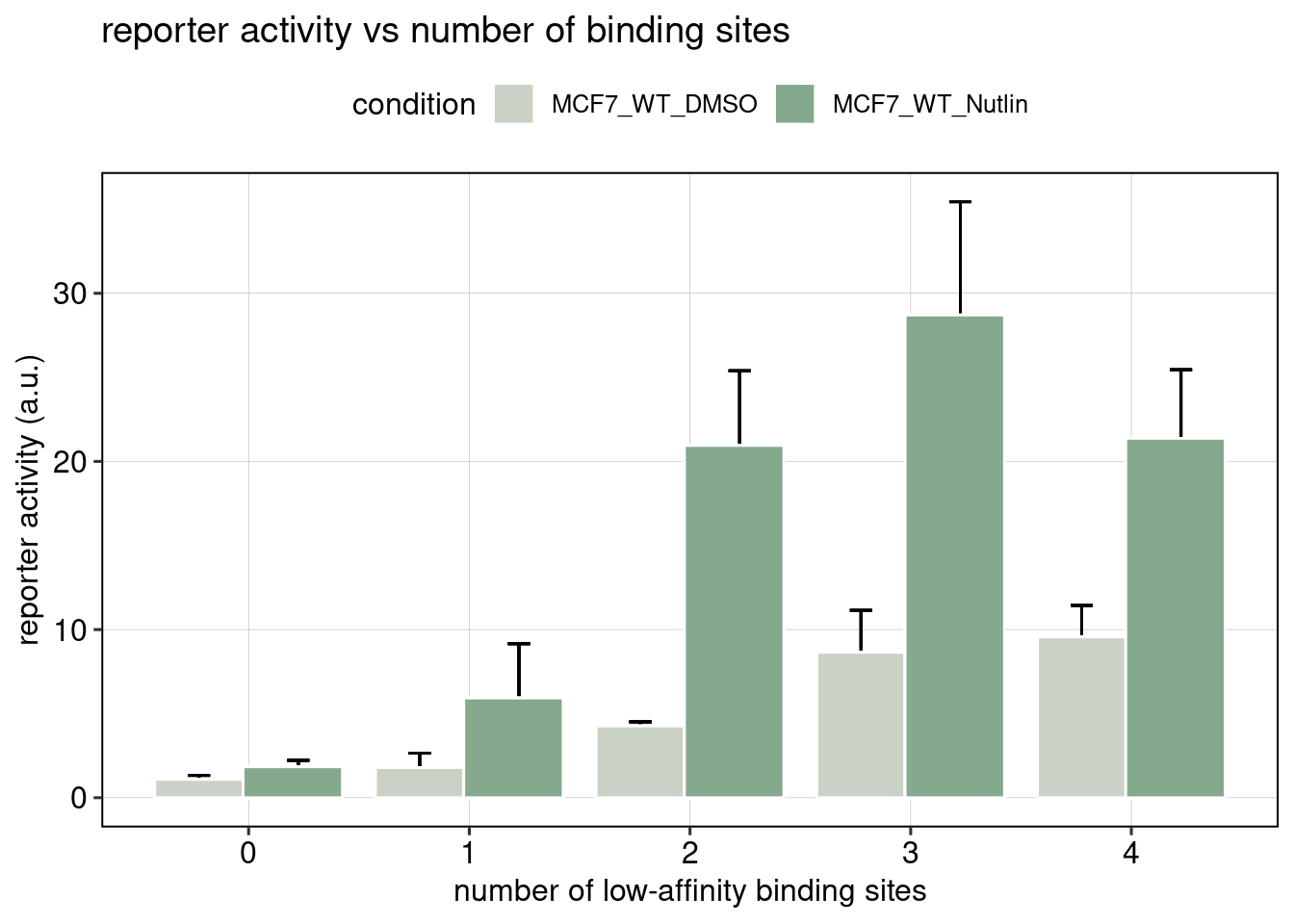
## [1] 0.0005569714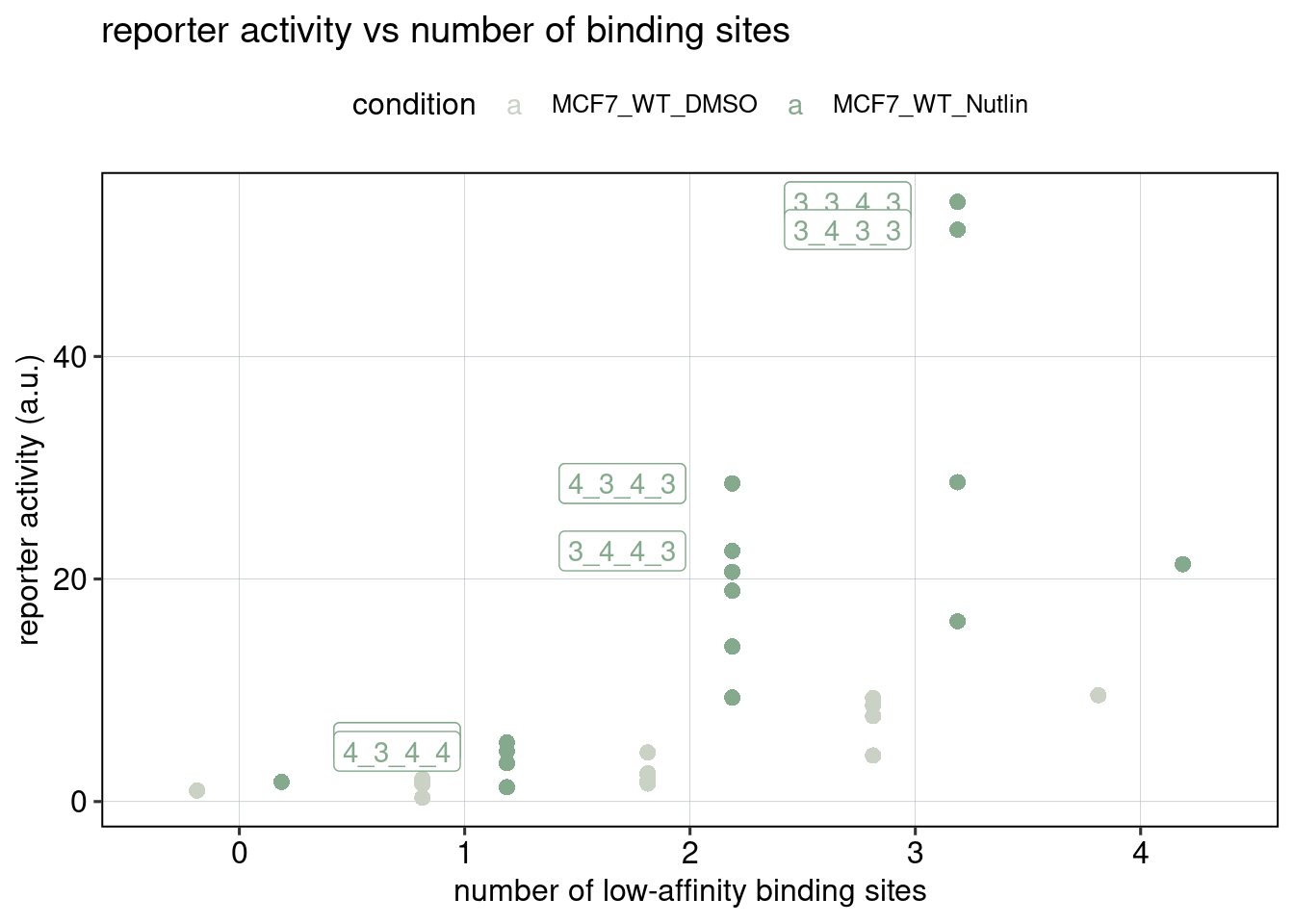
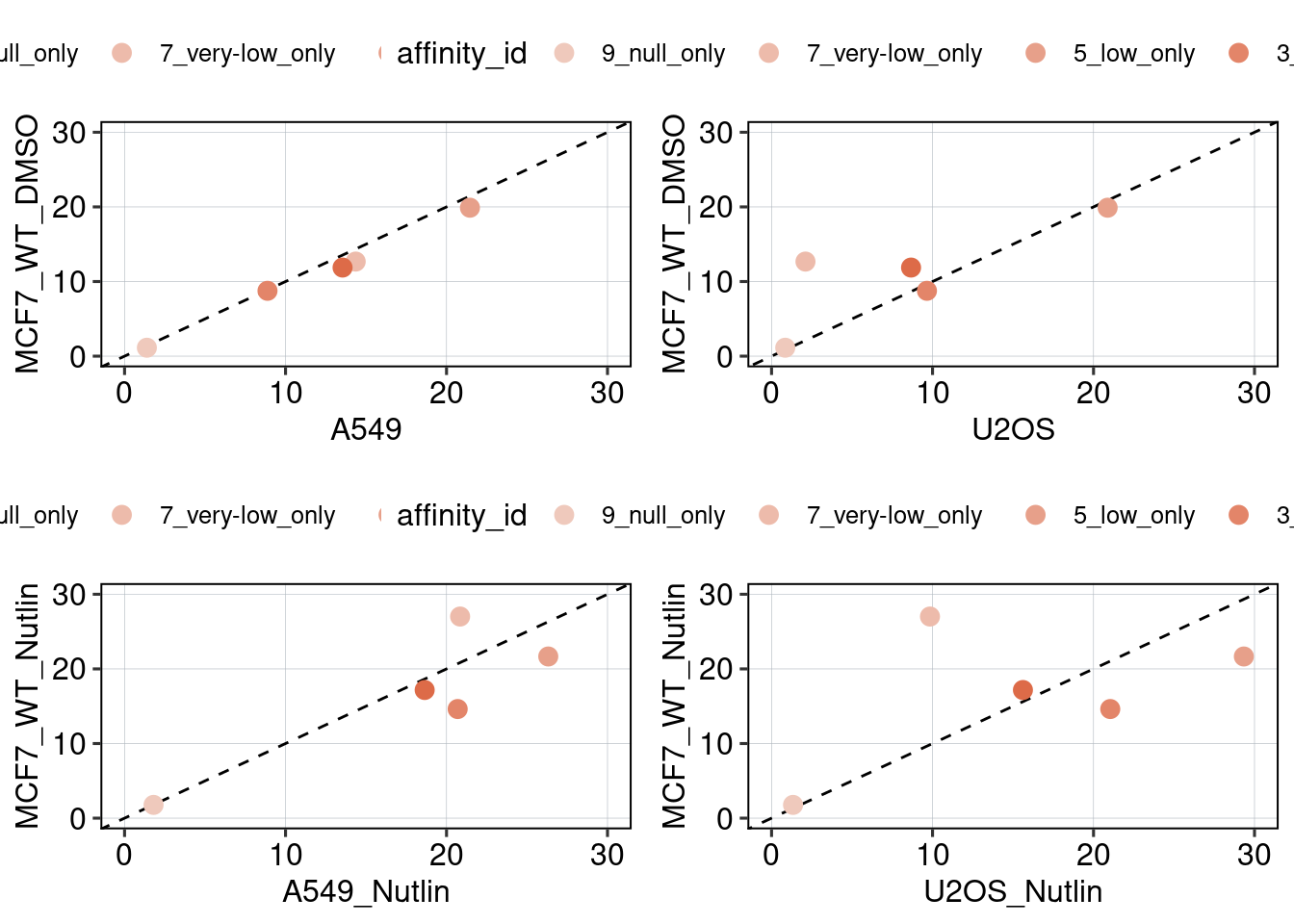
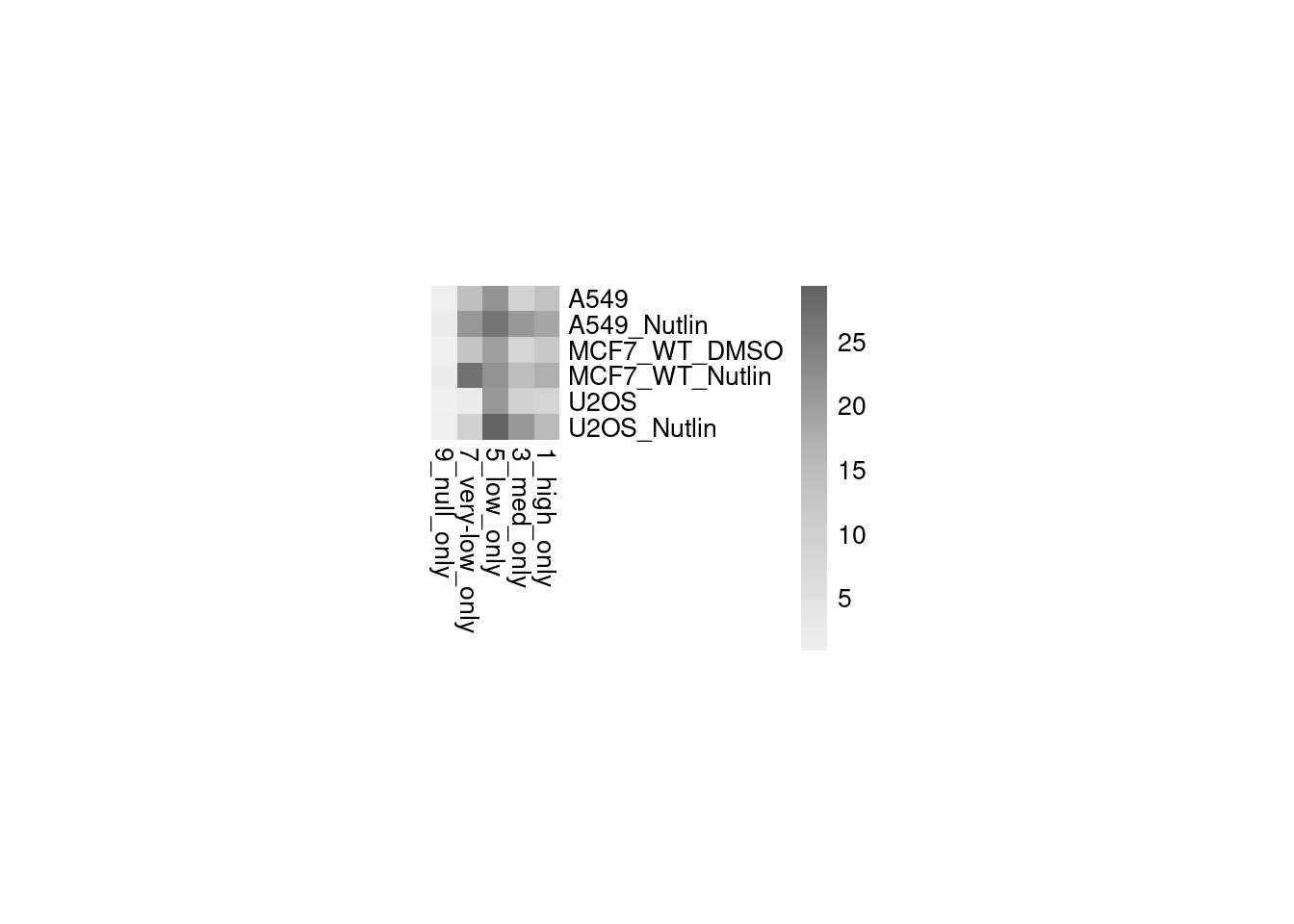
Conclusion: BS006 is the most responsive to Nutlin-3a. Addition of binding sites is super-additive. Positioning of binding sites matters - putting them directly next to each other is inhibitory, and putting them close to the TSS leads to higher activity.
Figure 3: The effect of the spacer length.
Aim: Show how the spacer length between adjacent binding sites affects reporter activity.
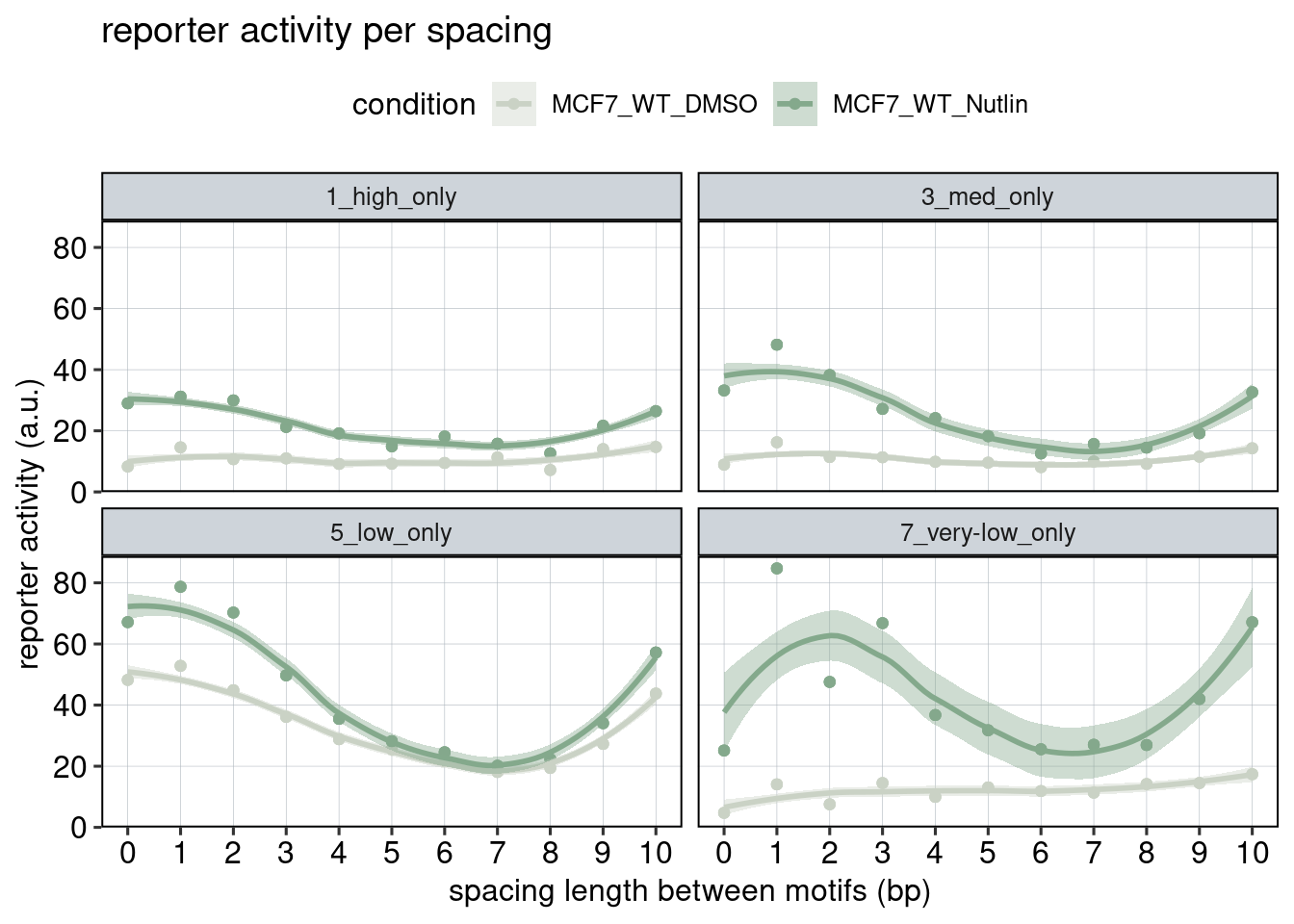
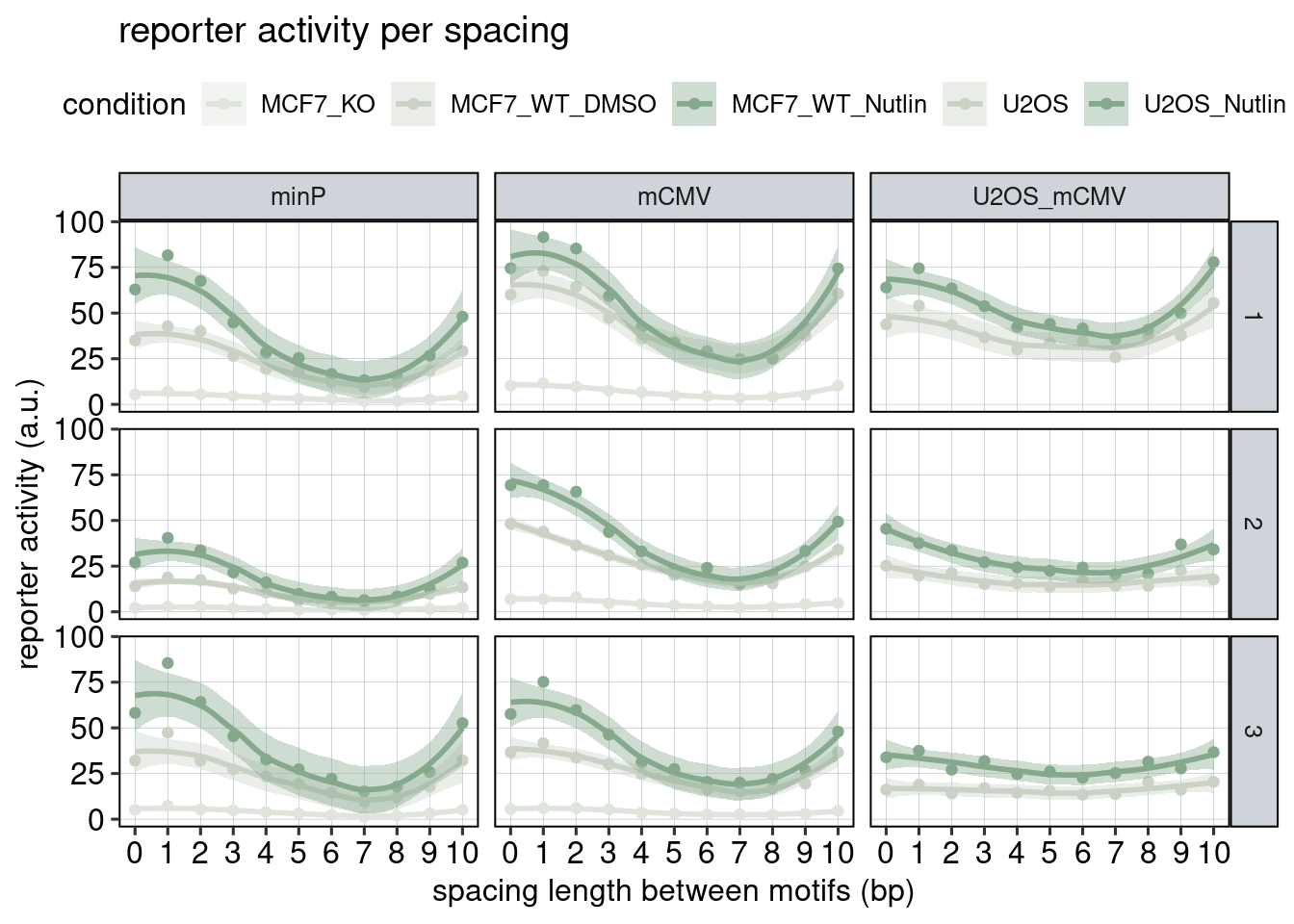
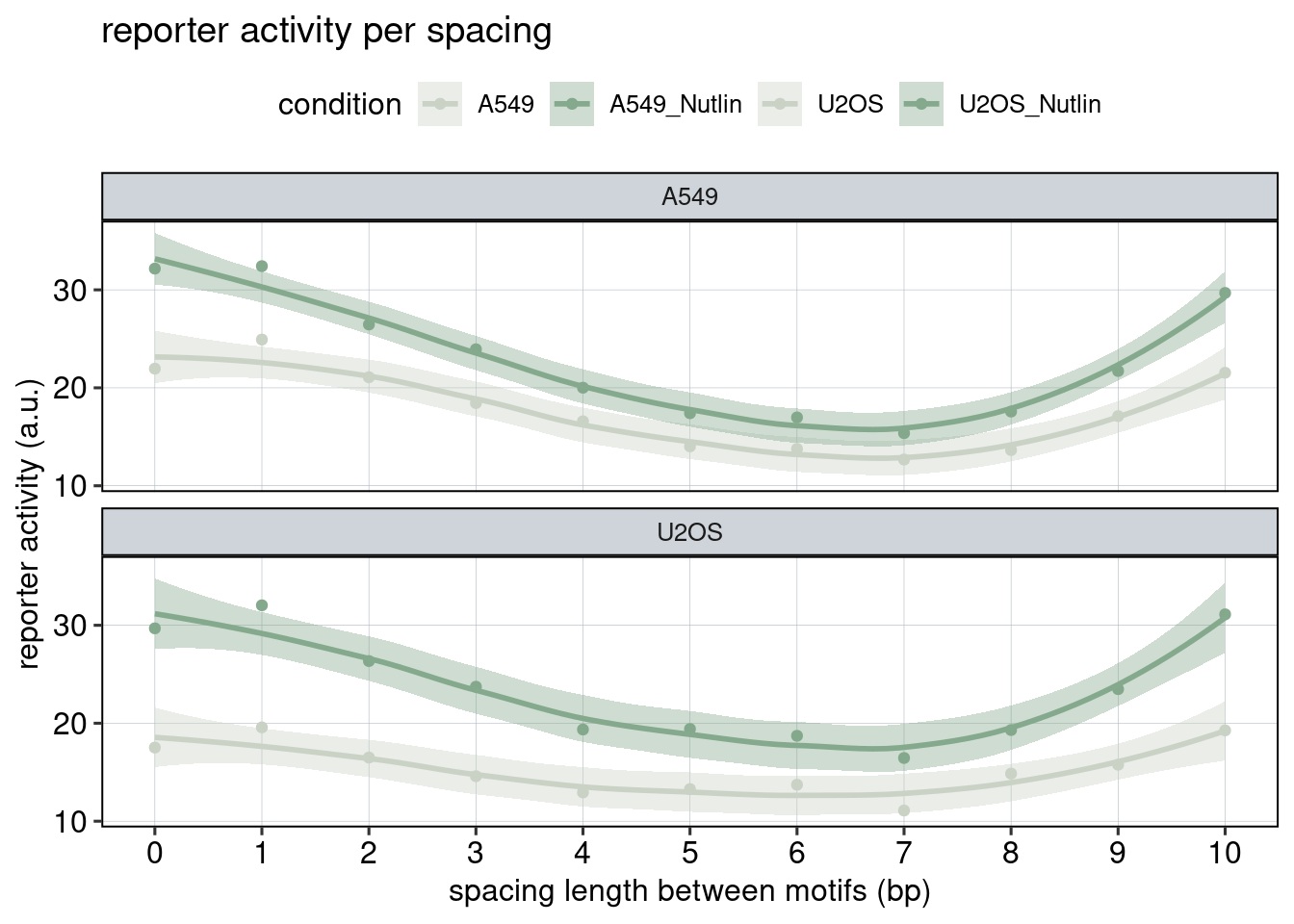
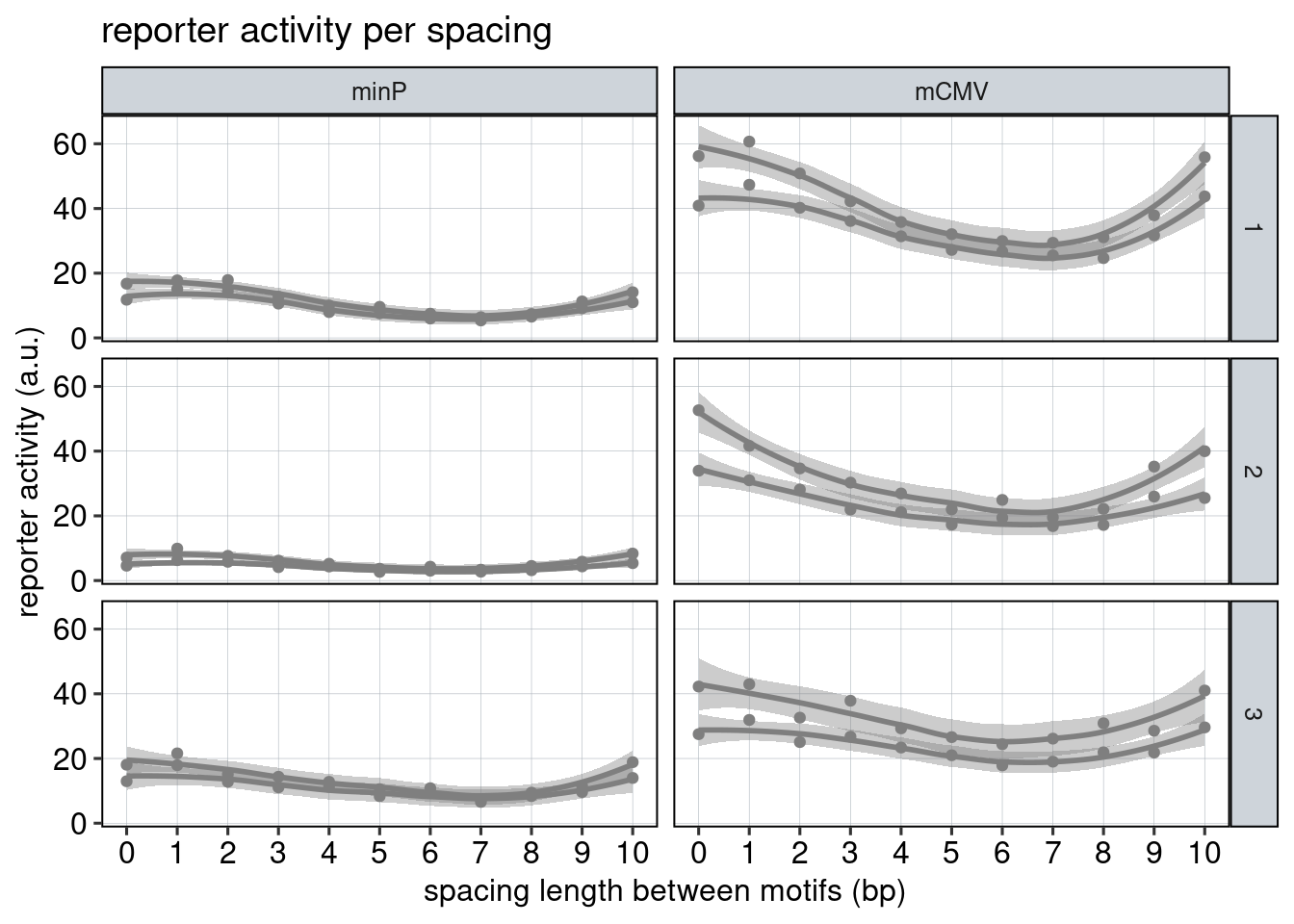
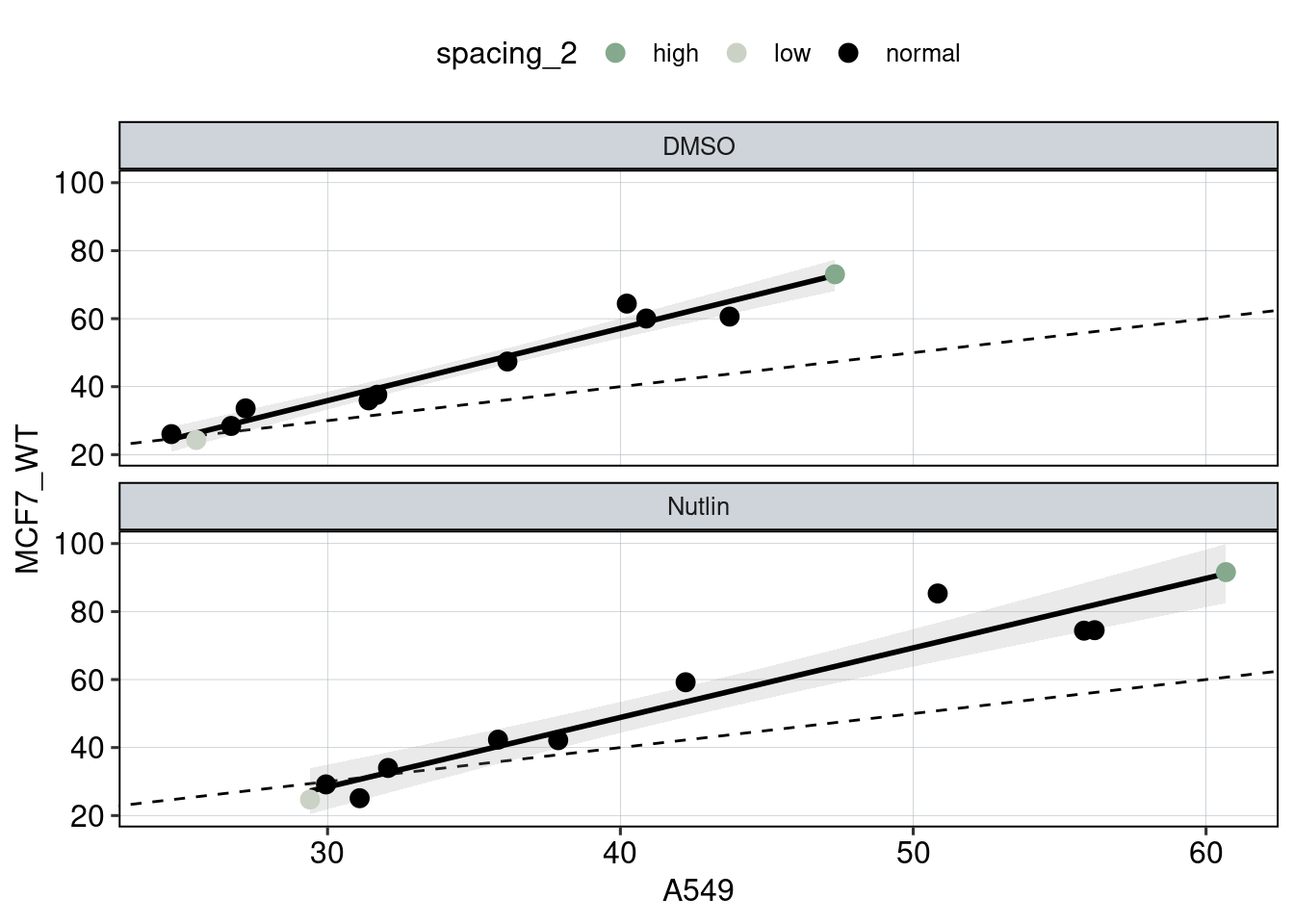
Conclusion: Spacer length influences activity periodically. Adjacent binding sites need to be 180 degrees tilted with respect to each other to achieve optimal activation.
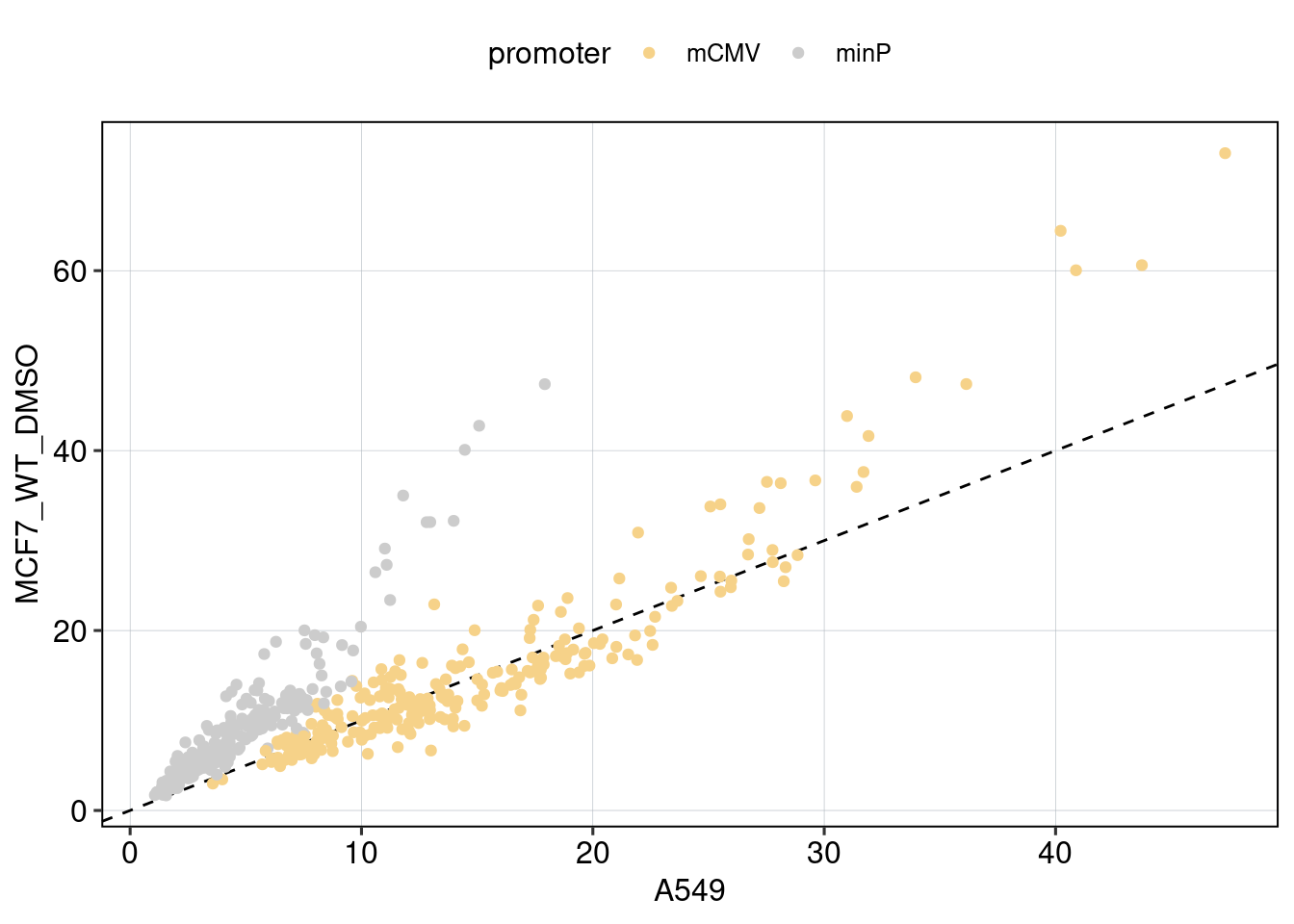
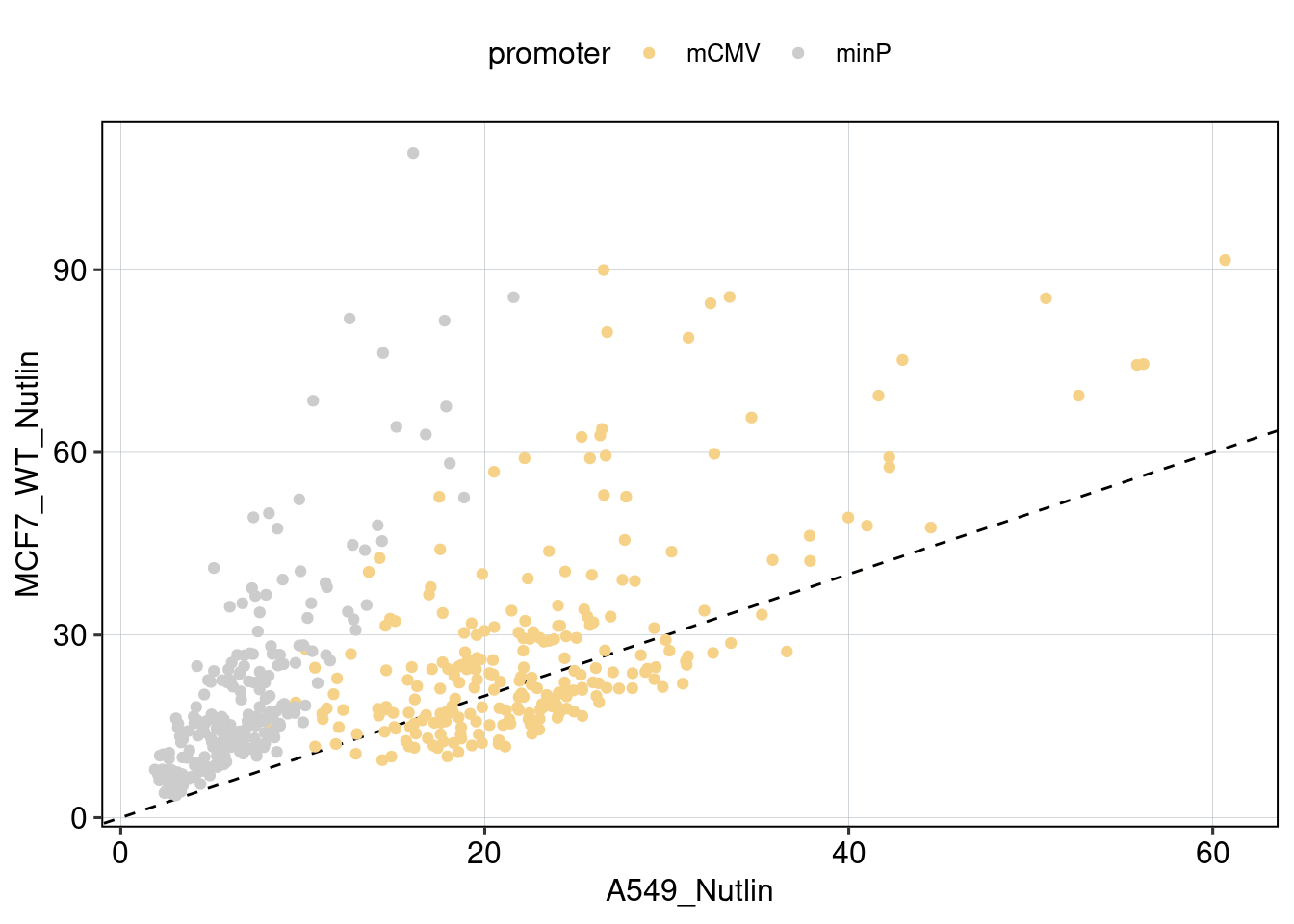
Figure 4: The effect of the minimal promoter and the spacer sequence.
Aim: Show how the P53 reporters interact with the two minimal promoters and the three spacer sequences.
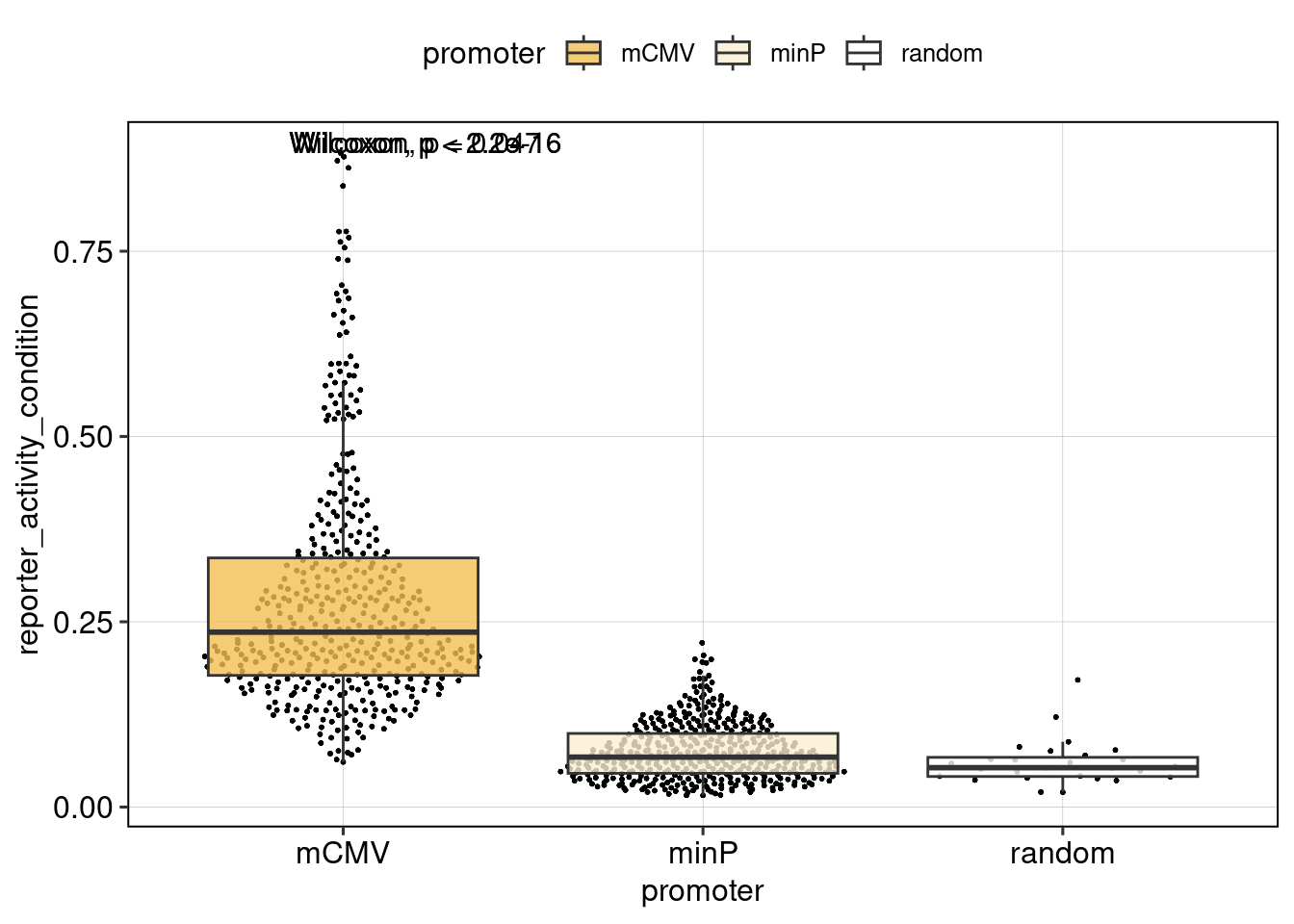
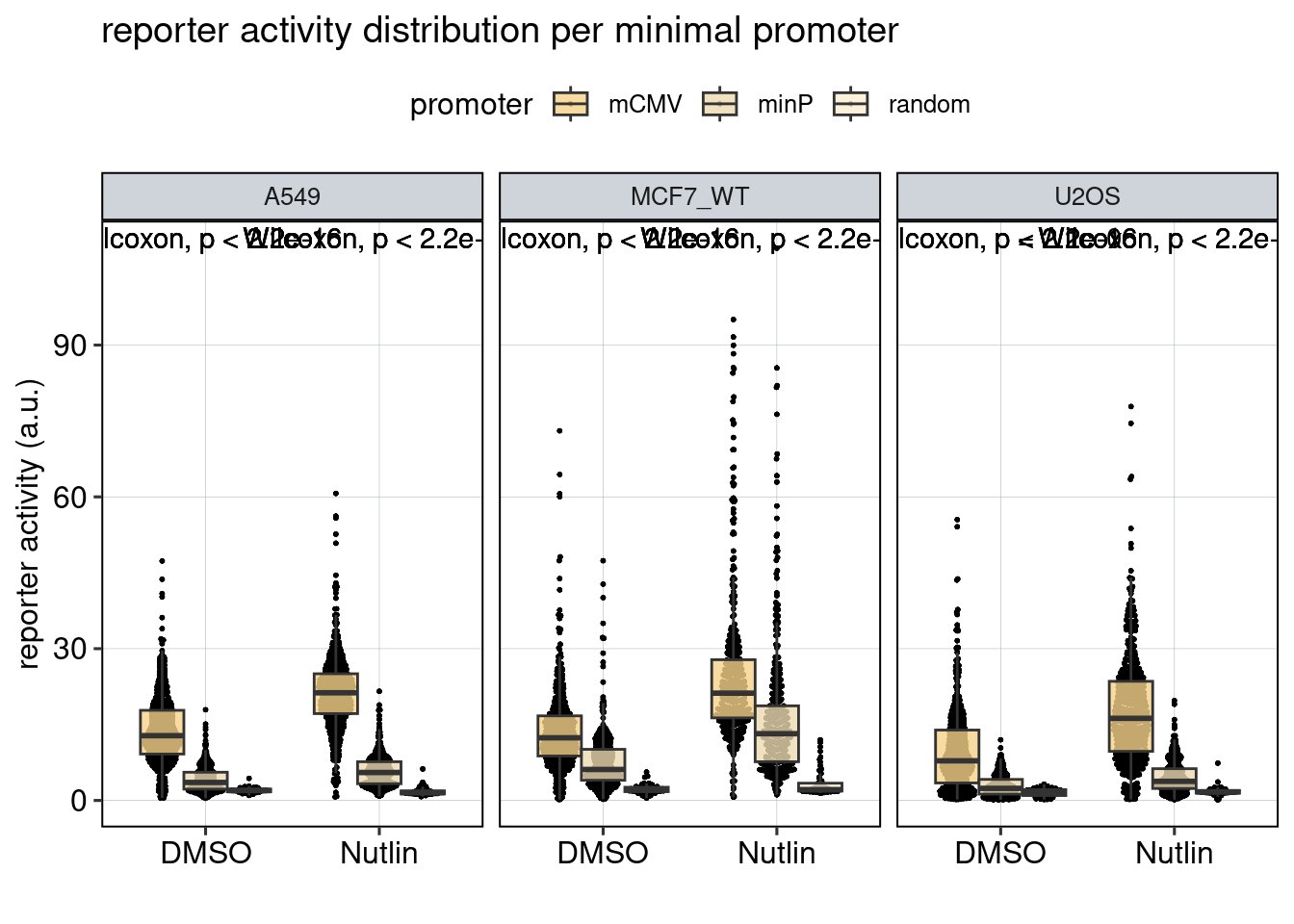
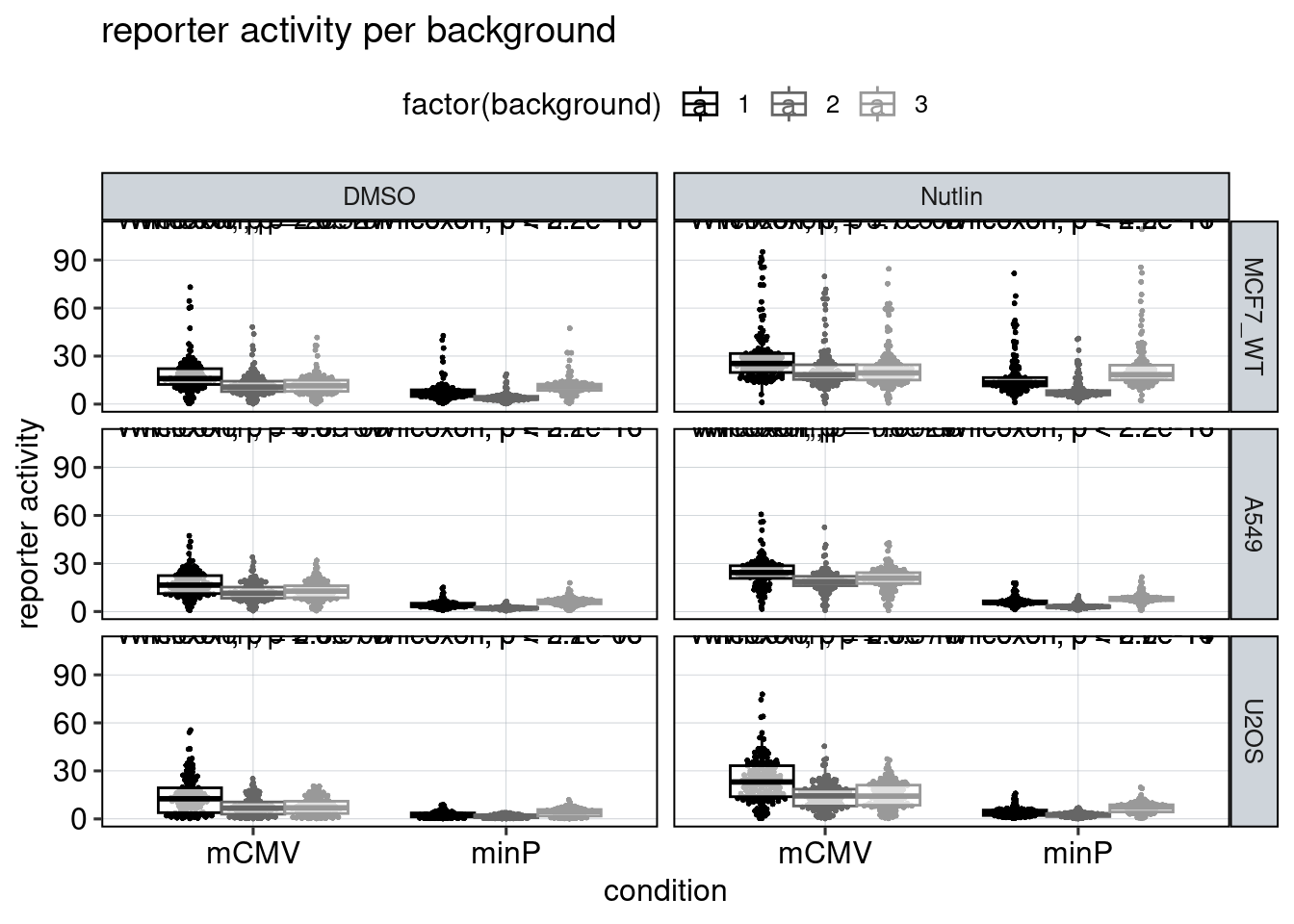
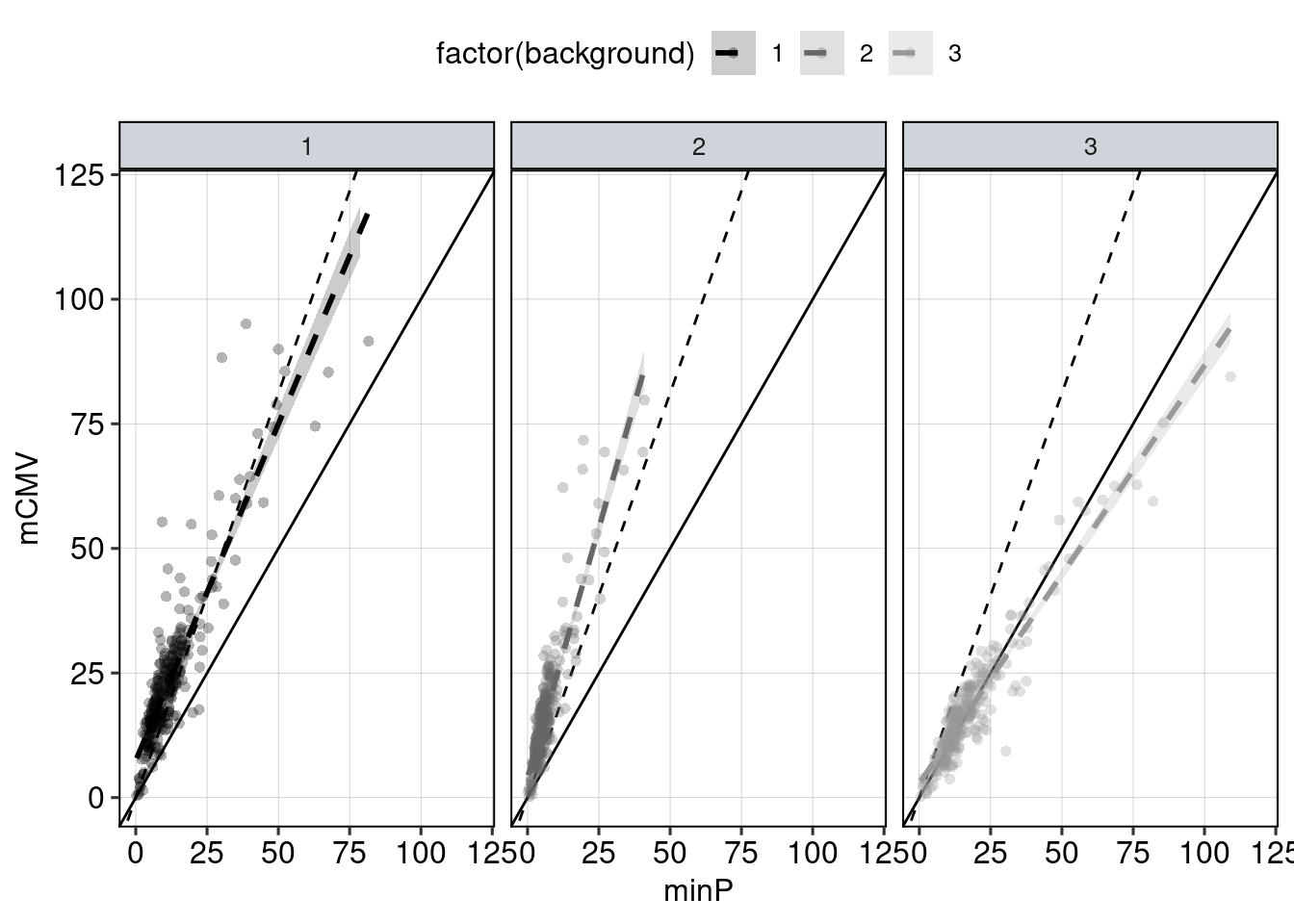
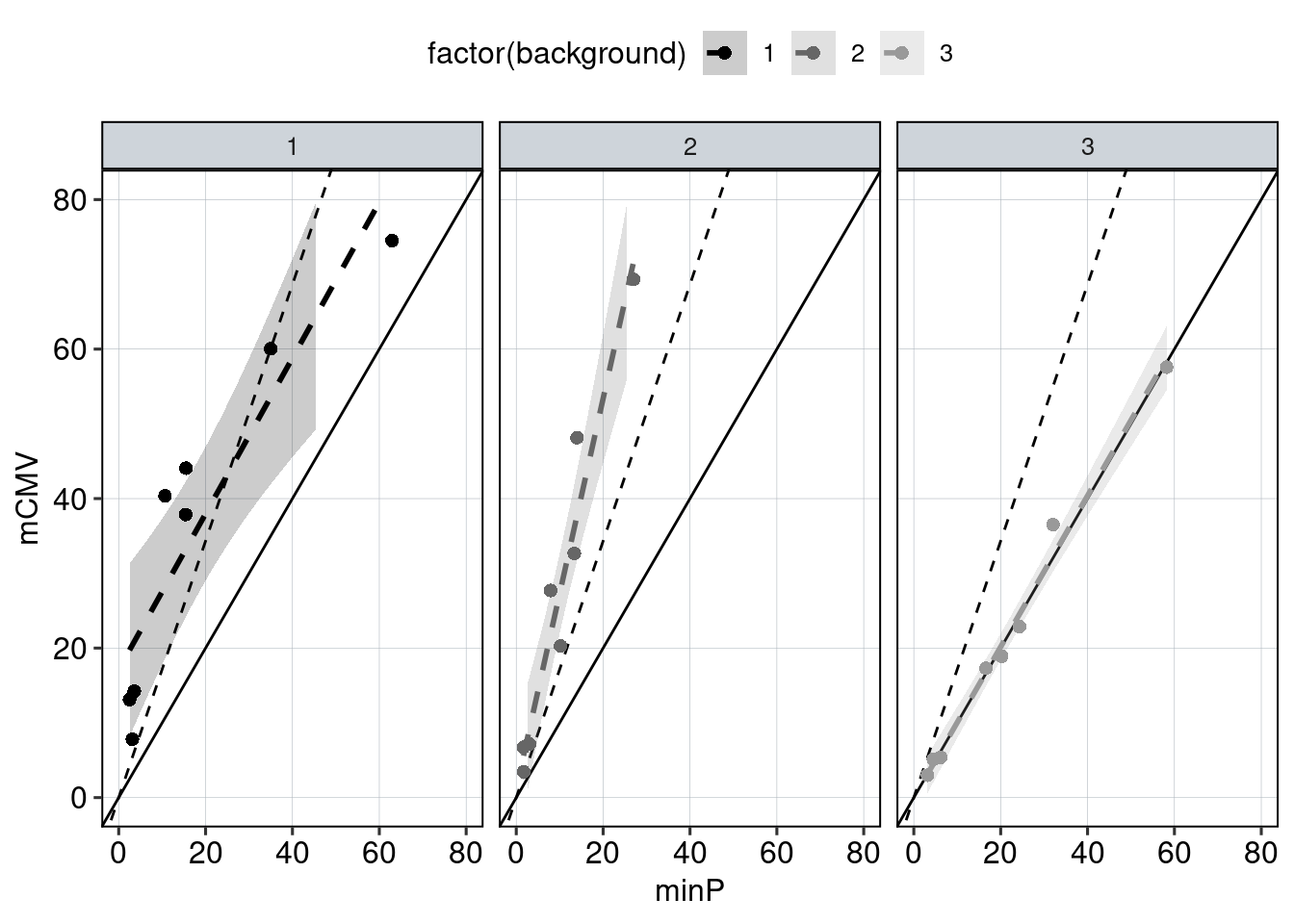
Conclusion: Promoter and spacer sequence influence activity linearly.
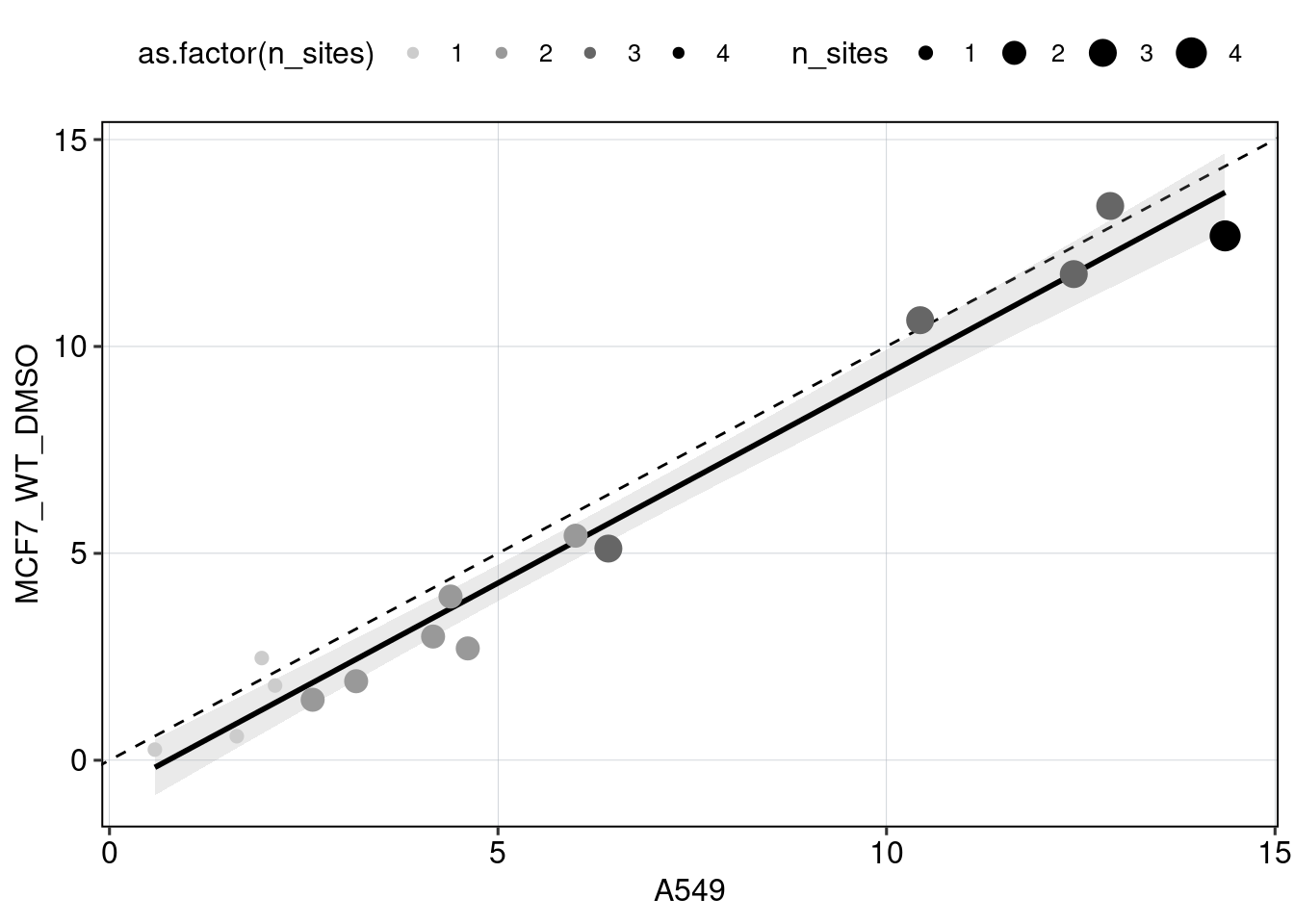
Figure 5 & 6: Linear model + Selection of best reporters
Aim: Can we explain now every observation using a linear model?
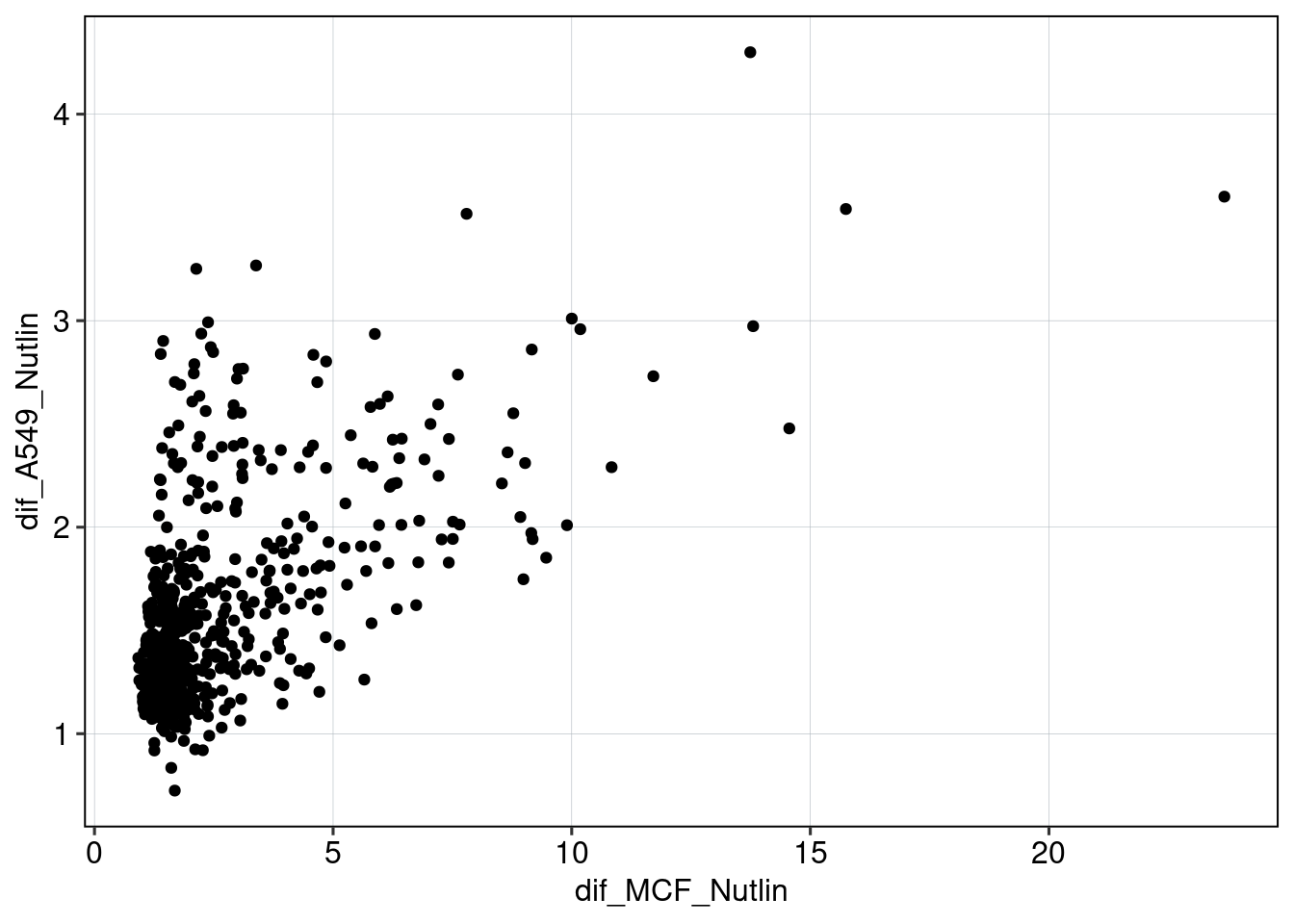
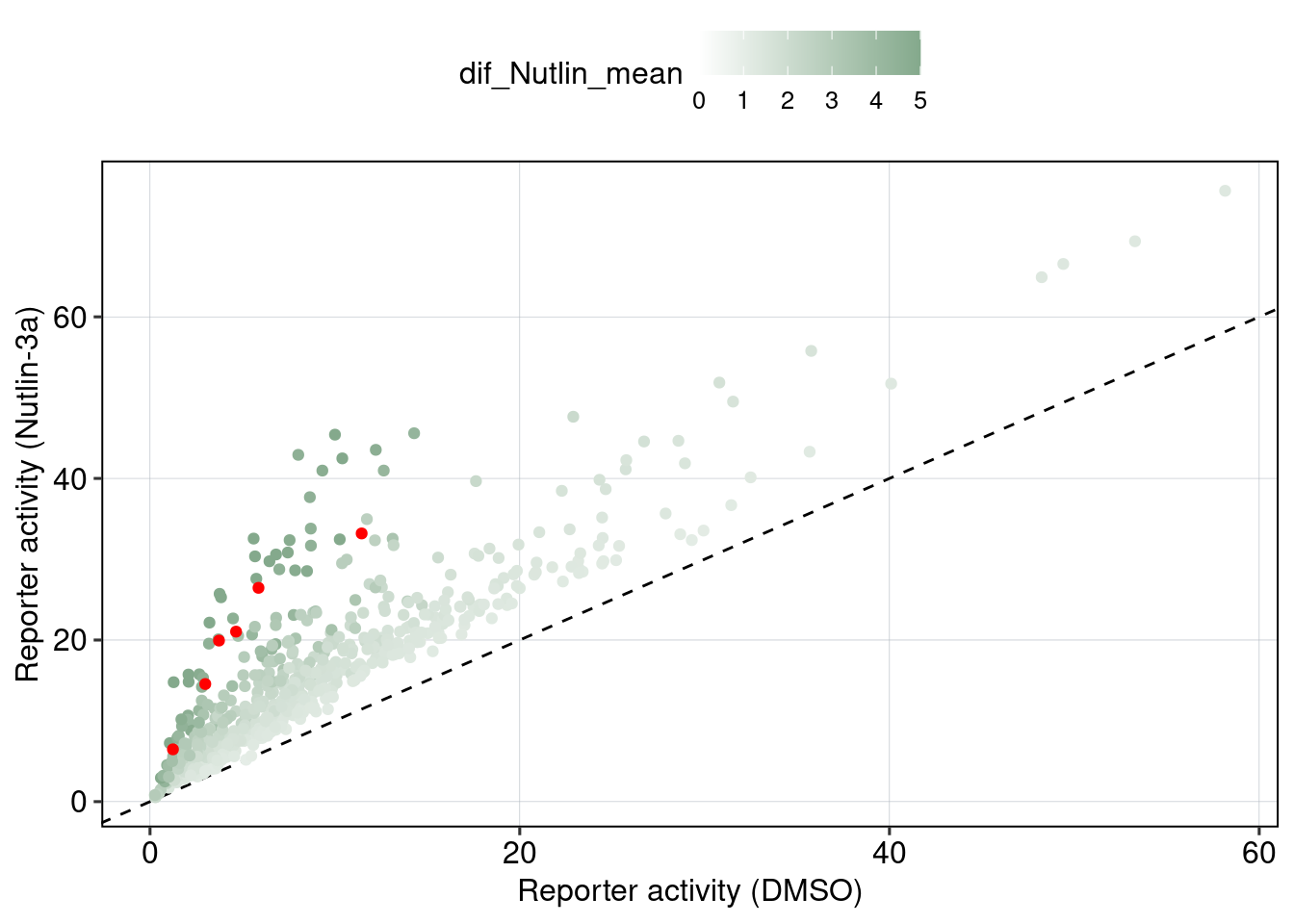
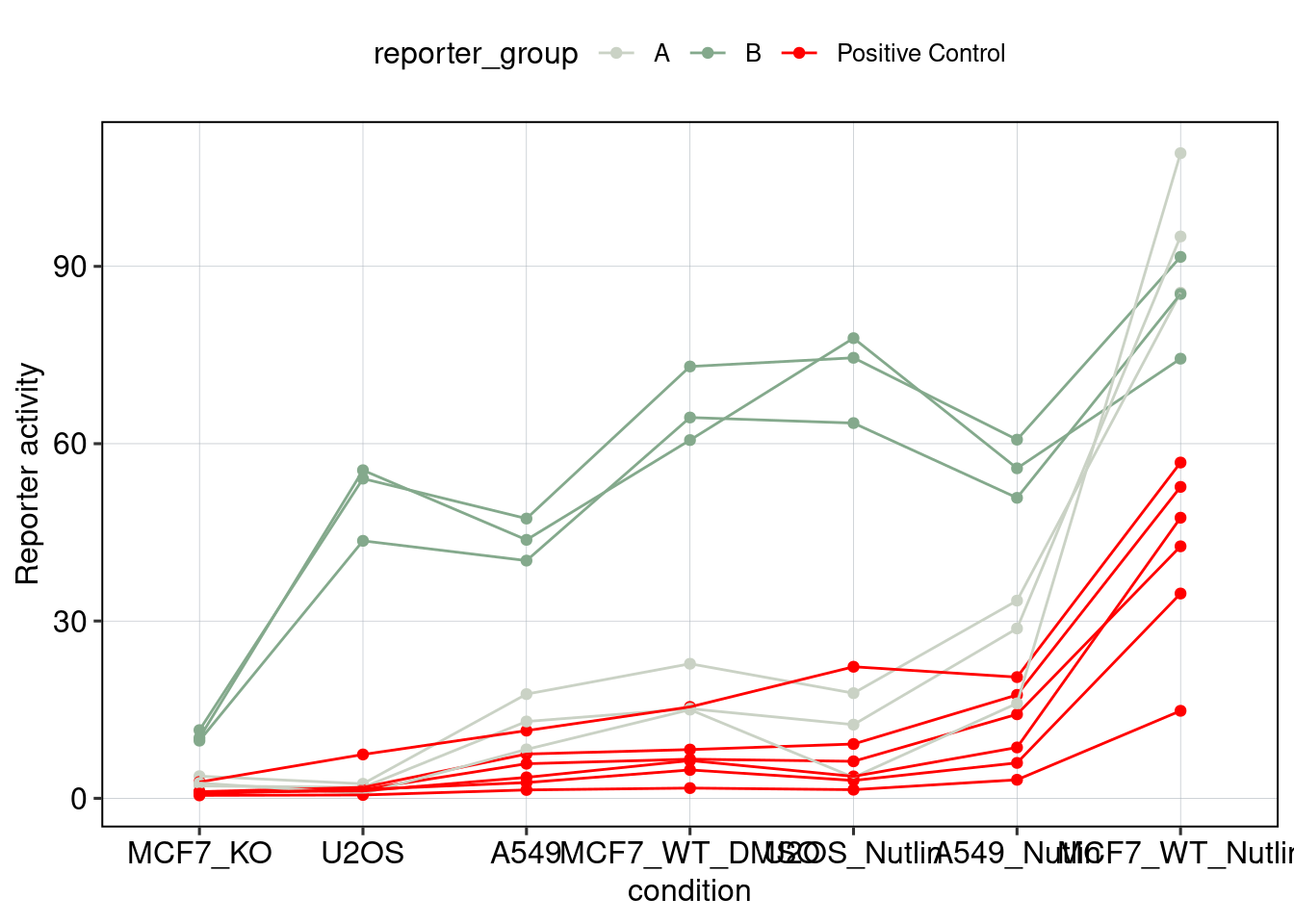
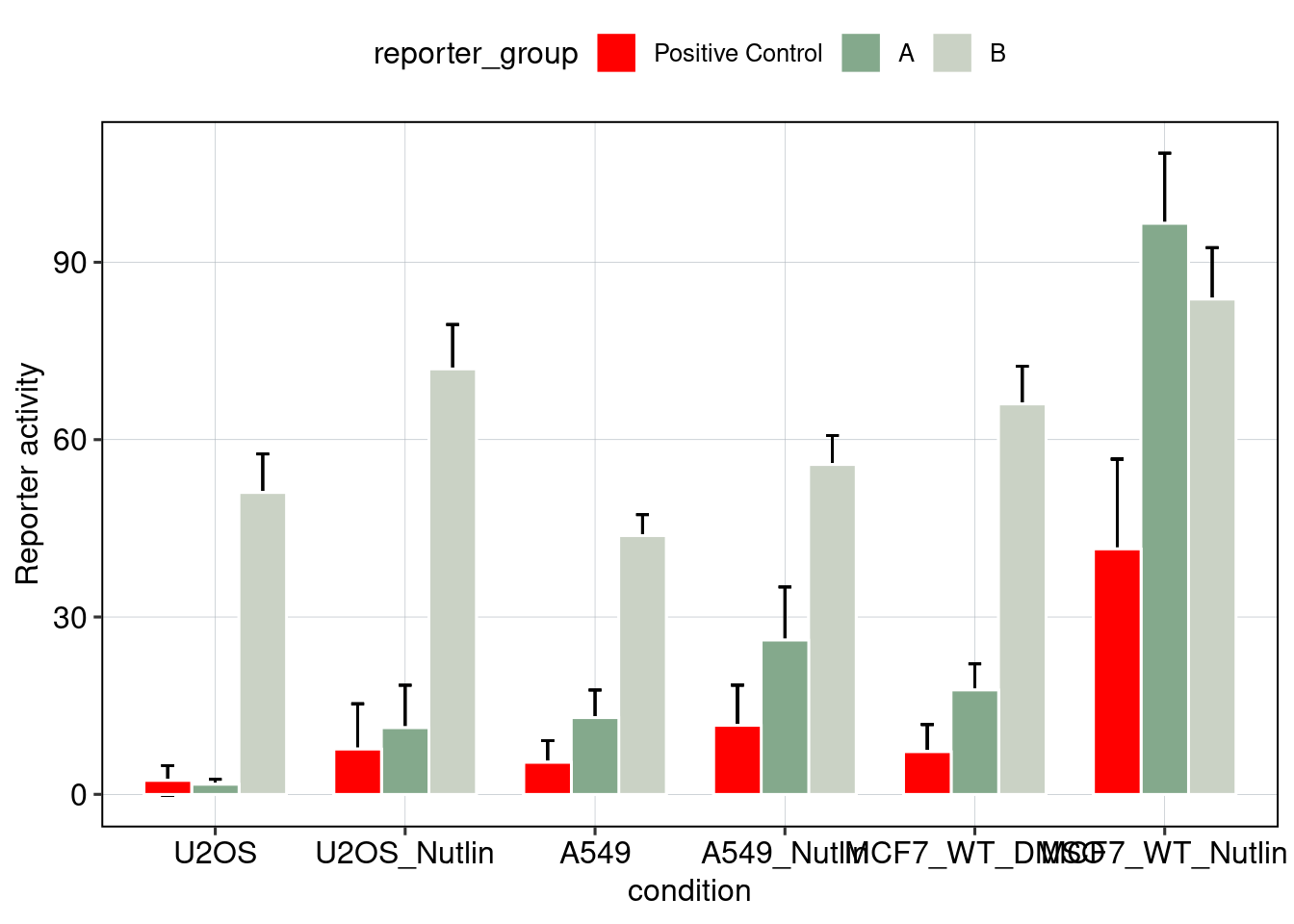
## [1] 0.08400584## MODEL INFO:
## Observations: 263 (1 missing obs. deleted)
## Dependent Variable: log2(reporter_activity)
## Type: OLS linear regression
##
## MODEL FIT:
## F(9,253) = 145.09, p = 0.00
## R² = 0.84
## Adj. R² = 0.83
##
## Standard errors: OLS
## ---------------------------------------------------------------
## Est. S.E. t val. p
## -------------------------------- ------- ------ -------- ------
## (Intercept) 3.07 0.07 41.69 0.00
## promotermCMV 1.30 0.08 15.39 0.00
## background2 -0.89 0.08 -10.59 0.00
## background3 0.37 0.08 4.45 0.00
## spacing_degree_transf 0.50 0.03 14.65 0.00
## affinity_id3_med_only 0.35 0.07 5.12 0.00
## affinity_id5_low_only 1.06 0.07 15.49 0.00
## affinity_id7_very-low_only 0.48 0.07 7.03 0.00
## promotermCMV:background2 0.38 0.12 3.19 0.00
## promotermCMV:background3 -0.82 0.12 -6.95 0.00
## ---------------------------------------------------------------
## MODEL INFO:
## Observations: 259 (5 missing obs. deleted)
## Dependent Variable: log2(reporter_activity)
## Type: OLS linear regression
##
## MODEL FIT:
## F(9,249) = 158.00, p = 0.00
## R² = 0.85
## Adj. R² = 0.85
##
## Standard errors: OLS
## ---------------------------------------------------------------
## Est. S.E. t val. p
## -------------------------------- ------- ------ -------- ------
## (Intercept) 2.09 0.09 24.50 0.00
## promotermCMV 1.60 0.10 16.73 0.00
## background2 -0.88 0.10 -9.15 0.00
## background3 0.53 0.10 5.49 0.00
## spacing_degree_transf 0.19 0.04 4.84 0.00
## affinity_id3_med_only -0.04 0.08 -0.52 0.60
## affinity_id5_low_only 1.41 0.08 17.97 0.00
## affinity_id7_very-low_only -0.26 0.08 -3.32 0.00
## promotermCMV:background2 0.19 0.14 1.42 0.16
## promotermCMV:background3 -1.15 0.13 -8.53 0.00
## ---------------------------------------------------------------
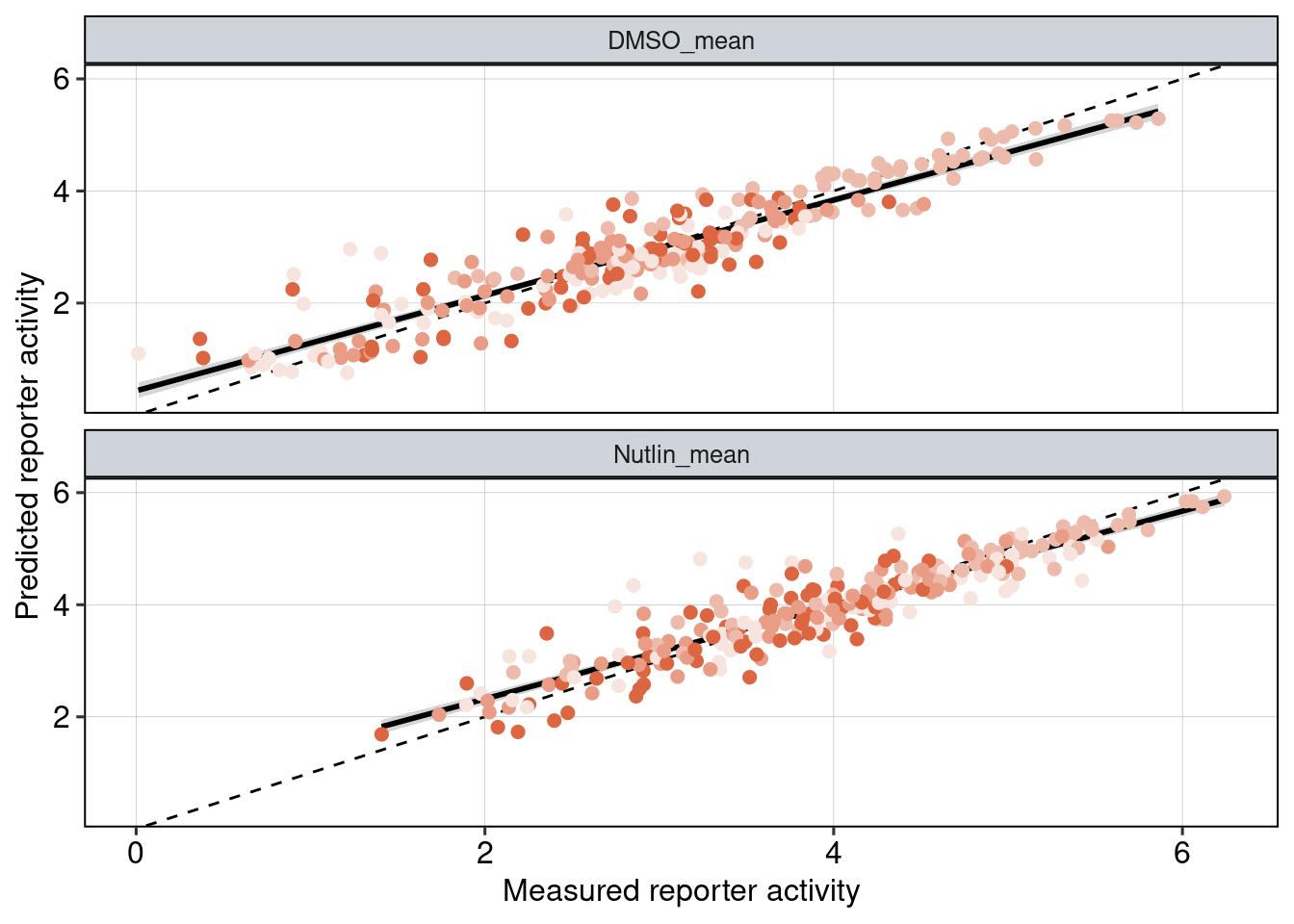
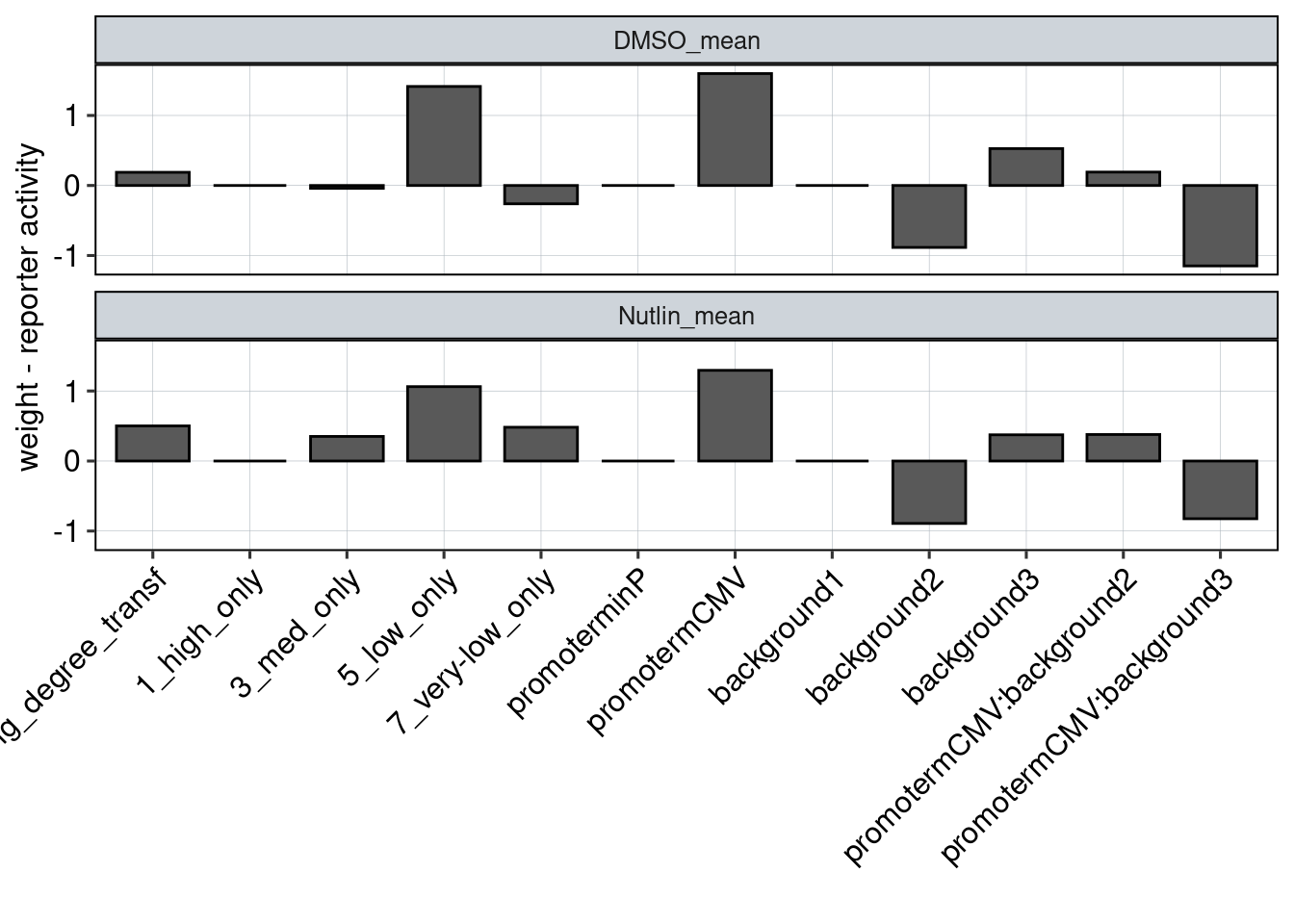
Conlusion: Top reporters are better than commercial reporters. Linear model gives insights into which features are important to drive high expression.
Session Info
paste("Run time: ",format(Sys.time()-StartTime))## [1] "Run time: 35.29388 secs"getwd()## [1] "/DATA/usr/m.trauernicht/projects/P53_reporter_scan/docs"date()## [1] "Wed Jun 14 09:42:36 2023"sessionInfo()## R version 4.0.5 (2021-03-31)
## Platform: x86_64-pc-linux-gnu (64-bit)
## Running under: Ubuntu 20.04.6 LTS
##
## Matrix products: default
## BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
## LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/liblapack.so.3
##
## locale:
## [1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8 LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8 LC_PAPER=en_US.UTF-8
## [8] LC_NAME=C LC_ADDRESS=C LC_TELEPHONE=C LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
##
## attached base packages:
## [1] stats4 grid parallel stats graphics grDevices utils datasets methods base
##
## other attached packages:
## [1] ggrastr_1.0.1 jtools_2.1.4 glmnetUtils_1.1.8 glmnet_4.1-4 Matrix_1.5-1 randomForest_4.6-14
## [7] ROCR_1.0-11 cowplot_1.1.1 ggforce_0.3.3 maditr_0.8.3 PCAtools_2.2.0 ggrepel_0.9.1
## [13] DESeq2_1.30.1 SummarizedExperiment_1.20.0 Biobase_2.50.0 MatrixGenerics_1.2.1 matrixStats_0.62.0 GenomicRanges_1.42.0
## [19] GenomeInfoDb_1.26.7 IRanges_2.24.1 S4Vectors_0.28.1 BiocGenerics_0.36.1 tidyr_1.2.0 viridis_0.6.2
## [25] viridisLite_0.4.0 ggpointdensity_0.1.0 ggbiplot_0.55 scales_1.2.0 factoextra_1.0.7.999 shiny_1.7.1
## [31] pheatmap_1.0.12 gridExtra_2.3 RColorBrewer_1.1-3 readr_2.1.2 haven_2.5.0 ggbeeswarm_0.6.0
## [37] plotly_4.10.0 tibble_3.1.6 dplyr_1.0.8 vwr_0.3.0 latticeExtra_0.6-29 lattice_0.20-41
## [43] stringdist_0.9.8 GGally_2.1.2 ggpubr_0.4.0 ggplot2_3.4.0 stringr_1.4.0 plyr_1.8.7
## [49] data.table_1.14.2
##
## loaded via a namespace (and not attached):
## [1] backports_1.4.1 lazyeval_0.2.2 splines_4.0.5 crosstalk_1.2.0 BiocParallel_1.24.1 digest_0.6.29 foreach_1.5.2
## [8] htmltools_0.5.2 fansi_1.0.3 magrittr_2.0.3 memoise_2.0.1 tzdb_0.3.0 annotate_1.68.0 vroom_1.5.7
## [15] prettyunits_1.1.1 jpeg_0.1-9 colorspace_2.0-3 blob_1.2.3 gitcreds_0.1.1 xfun_0.30 crayon_1.5.1
## [22] RCurl_1.98-1.6 jsonlite_1.8.0 genefilter_1.72.1 iterators_1.0.14 survival_3.2-10 glue_1.6.2 polyclip_1.10-0
## [29] gtable_0.3.0 zlibbioc_1.36.0 XVector_0.30.0 DelayedArray_0.16.3 car_3.0-12 BiocSingular_1.6.0 shape_1.4.6
## [36] abind_1.4-5 DBI_1.1.2 rstatix_0.7.0 Rcpp_1.0.8.3 progress_1.2.2 xtable_1.8-4 dqrng_0.3.0
## [43] bit_4.0.4 rsvd_1.0.5 htmlwidgets_1.5.4 httr_1.4.2 ellipsis_0.3.2 farver_2.1.0 pkgconfig_2.0.3
## [50] reshape_0.8.9 XML_3.99-0.9 sass_0.4.1 locfit_1.5-9.4 utf8_1.2.2 labeling_0.4.2 tidyselect_1.1.2
## [57] rlang_1.0.6 reshape2_1.4.4 later_1.3.0 AnnotationDbi_1.52.0 munsell_0.5.0 tools_4.0.5 cachem_1.0.6
## [64] cli_3.4.1 generics_0.1.2 RSQLite_2.2.12 broom_0.8.0 evaluate_0.15 fastmap_1.1.0 yaml_2.3.5
## [71] knitr_1.38 bit64_4.0.5 pander_0.6.5 purrr_0.3.4 nlme_3.1-152 sparseMatrixStats_1.2.1 mime_0.12
## [78] compiler_4.0.5 rstudioapi_0.13 beeswarm_0.4.0 png_0.1-7 ggsignif_0.6.3 tweenr_1.0.2 geneplotter_1.68.0
## [85] bslib_0.3.1 stringi_1.7.6 highr_0.9 forcats_0.5.1 vctrs_0.5.1 pillar_1.7.0 lifecycle_1.0.3
## [92] jquerylib_0.1.4 bitops_1.0-7 irlba_2.3.5 httpuv_1.6.5 R6_2.5.1 promises_1.2.0.1 vipor_0.4.5
## [99] codetools_0.2-18 MASS_7.3-53.1 assertthat_0.2.1 withr_2.5.0 GenomeInfoDbData_1.2.4 mgcv_1.8-34 hms_1.1.1
## [106] beachmat_2.6.4 rmarkdown_2.13 DelayedMatrixStats_1.12.3 carData_3.0-5 Cairo_1.5-15